A transcriptionally active subgenomic promoter supports homologous crossovers in a plus-strand RNA virus
- PMID: 12767997
- PMCID: PMC156210
- DOI: 10.1128/jvi.77.12.6769-6776.2003
A transcriptionally active subgenomic promoter supports homologous crossovers in a plus-strand RNA virus
Abstract
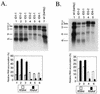
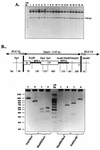
Genetic RNA recombination plays an important role in viral evolution, but its molecular mechanism is not well understood. In this work we describe homologous RNA recombination activity that is supported by a subgenomic promoter (sgp) region in the RNA3 segment of brome mosaic bromovirus (BMV), a tripartite plus-strand RNA virus. The crossover frequencies were determined by coinoculations with pairs of BMV RNA3 variants that carried a duplicated sgp region flanked by marker restriction sites. A region composed of the sgp core, a poly(A) tract, and an upstream enhancer supported homologous exchanges in 25% of the analyzed RNA3 progeny. However, mutations in the sgp core stopped both the transcription of the sgp RNA and homologous recombination. These data provide evidence for an association of RNA recombination with transcription.
Figures


Similar articles
-
Dissecting the requirement for subgenomic promoter sequences by RNA recombination of brome mosaic virus in vivo: evidence for functional separation of transcription and recombination.J Virol. 2004 Aug;78(16):8552-64. doi: 10.1128/JVI.78.16.8552-8564.2004. J Virol. 2004. PMID: 15280464 Free PMC article.
-
Recombination of 5' subgenomic RNA3a with genomic RNA3 of Brome mosaic bromovirus in vitro and in vivo.Virology. 2011 Feb 5;410(1):129-41. doi: 10.1016/j.virol.2010.10.037. Epub 2010 Nov 26. Virology. 2011. PMID: 21111438 Free PMC article.
-
Homologous crossovers among molecules of brome mosaic bromovirus RNA1 or RNA2 segments in vivo.J Virol. 2005 May;79(9):5732-42. doi: 10.1128/JVI.79.9.5732-5742.2005. J Virol. 2005. PMID: 15827188 Free PMC article.
-
RNA recombination in brome mosaic virus, a model plus strand RNA virus.Acta Biochim Pol. 1998;45(4):847-68. Acta Biochim Pol. 1998. PMID: 10397334 Review.
-
The brome mosaic virus 3' untranslated sequence regulates RNA replication, recombination, and virion assembly.Virus Res. 2015 Aug 3;206:46-52. doi: 10.1016/j.virusres.2015.02.007. Epub 2015 Feb 14. Virus Res. 2015. PMID: 25687214 Review.
Cited by
-
The AU-rich RNA recombination hot spot sequence of Brome mosaic virus is functional in tombusviruses: implications for the mechanism of RNA recombination.J Virol. 2004 Mar;78(5):2288-300. doi: 10.1128/jvi.78.5.2288-2300.2004. J Virol. 2004. PMID: 14963125 Free PMC article.
-
Defective Interfering RNAs: Foes of Viruses and Friends of Virologists.Viruses. 2009 Dec;1(3):895-919. doi: 10.3390/v1030895. Epub 2009 Nov 10. Viruses. 2009. PMID: 21994575 Free PMC article.
-
Long Noncoding RNAs in Plant Viroids and Viruses: A Review.Pathogens. 2020 Sep 18;9(9):765. doi: 10.3390/pathogens9090765. Pathogens. 2020. PMID: 32961969 Free PMC article. Review.
-
Phylogenetic and Molecular Variability Studies Reveal a New Genetic Clade of Citrus leprosis virus C.Viruses. 2016 Jun 6;8(6):153. doi: 10.3390/v8060153. Viruses. 2016. PMID: 27275832 Free PMC article.
-
Co-infection with two strains of Brome mosaic bromovirus reveals common RNA recombination sites in different hosts.Virus Evol. 2015 Dec 23;1(1):vev021. doi: 10.1093/ve/vev021. eCollection 2015. Virus Evol. 2015. PMID: 27774290 Free PMC article.
References
-
- Adkins, S., and C. C. Kao. 1998. Subgenomic RNA promoters dictate the mode of recognition by bromoviral RNA-dependent RNA polymerases. Virology 252:1-8. - PubMed
-
- Ahlquist, P. 1992. Bromovirus RNA replication and transcription. Curr. Opin. Genet. Dev. 2:71-76. - PubMed
-
- Balakrishnan, M., P. J. Fay, and R. A. Bambara. 2001. The kissing hairpin sequence promotes recombination within the HIV-I 5′ leader region. J. Biol. Chem. 276:36482-36492. - PubMed
-
- Bar-Joseph, M., G. Yang, R. Gafny, and M. Mawass. 1997. Subgenomic RNAs: the possible building blocks for modular recombination of Closteroviridae genomes. Semin. Virol. 8:113-119.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

