Karyotype analysis of four Vicia species using in situ hybridization with repetitive sequences
- PMID: 12770847
- PMCID: PMC4242401
- DOI: 10.1093/aob/mcg099
Karyotype analysis of four Vicia species using in situ hybridization with repetitive sequences
Abstract
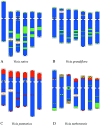
Mitotic chromosomes of four Vicia species (V. sativa, V. grandiflora, V. pannonica and V. narbonensis) were subjected to in situ hybridization with probes derived from conserved plant repetitive DNA sequences (18S-25S and 5S rDNA, telomeres) and genus-specific satellite repeats (VicTR-A and VicTR-B). Numbers and positions of hybridization signals provided cytogenetic landmarks suitable for unambiguous identification of all chromosomes, and establishment of the karyotypes. The VicTR-A and -B sequences, in particular, produced highly informative banding patterns that alone were sufficient for discrimination of all chromosomes. However, these patterns were not conserved among species and thus could not be employed for identification of homologous chromosomes. This fact, together with observed variations in positions and numbers of rDNA loci, suggests considerable divergence between karyotypes of the species studied.
Figures


Similar articles
-
Sequence homogenization and chromosomal localization of VicTR-B satellites differ between closely related Vicia species.Chromosoma. 2006 Dec;115(6):437-47. doi: 10.1007/s00412-006-0070-8. Epub 2006 Jun 20. Chromosoma. 2006. PMID: 16788823
-
Sequence subfamilies of satellite repeats related to rDNA intergenic spacer are differentially amplified on Vicia sativa chromosomes.Chromosoma. 2003 Oct;112(3):152-8. doi: 10.1007/s00412-003-0255-3. Epub 2003 Sep 27. Chromosoma. 2003. PMID: 14579131
-
Distribution of highly repeated DNA sequences in species of the genus Lens Miller.Genome. 2003 Dec;46(6):1118-24. doi: 10.1139/g03-077. Genome. 2003. PMID: 14663530
-
Microarray-based survey of repetitive genomic sequences in Vicia spp.Plant Mol Biol. 2001 Jan;45(2):229-44. doi: 10.1023/a:1006408119740. Plant Mol Biol. 2001. PMID: 11289513
-
Chromosome Banding in Amphibia. XXXII. The Genus Xenopus (Anura, Pipidae).Cytogenet Genome Res. 2015;145(3-4):201-17. doi: 10.1159/000433481. Epub 2015 Jun 25. Cytogenet Genome Res. 2015. PMID: 26112092 Review.
Cited by
-
Comparative mapping between Medicago sativa and Pisum sativum.Mol Genet Genomics. 2004 Oct;272(3):235-46. doi: 10.1007/s00438-004-1055-z. Epub 2004 Sep 1. Mol Genet Genomics. 2004. PMID: 15340836
-
Genome downsizing and karyotype constancy in diploid and polyploid congeners: a model of genome size variation.AoB Plants. 2014 Jun 26;6:plu029. doi: 10.1093/aobpla/plu029. AoB Plants. 2014. PMID: 24969503 Free PMC article.
-
Isolation and characterization of a new repetitive DNA family recently amplified in the Mesoamerican gene pool of the common bean (Phaseolus vulgaris L., Fabaceae).Genetica. 2011 Sep;139(9):1135-42. doi: 10.1007/s10709-011-9615-8. Epub 2011 Nov 16. Genetica. 2011. PMID: 22086374
-
Survey of extrachromosomal circular DNA derived from plant satellite repeats.BMC Plant Biol. 2008 Aug 22;8:90. doi: 10.1186/1471-2229-8-90. BMC Plant Biol. 2008. PMID: 18721471 Free PMC article.
-
Pod, seed traits and cytotaxonomic studies of some Vicia narbonensis L. accessions (Fabaceae).Saudi J Biol Sci. 2017 Nov;24(7):1689-1696. doi: 10.1016/j.sjbs.2015.11.003. Epub 2015 Nov 14. Saudi J Biol Sci. 2017. PMID: 30294236 Free PMC article.
References
-
- AllkinR, Goyder DJ, Bisby FA, White RJ.1986.Names and synonyms of species and subspecies in the Vicieae: Issue 3. Vicieae Database Project, Publication No 7 (ISSN 0263‐8517), University of Southampton, UK.
-
- BennettMD, Leitch IJ.1998. Angiosperm DNA C‐values database. http://www.rgbkew.org.uk/cval/homepage.html
-
- BuschW, Herrmann RG, Houben A, Martin R.1996. Efficient preparation of plant metaphase spreads. Plant Molecular Biology Reporter 14: 149–155.
-
- CharlesworthB, Sniegowski P, Stephan W.1994. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371: 215–220. - PubMed
-
- CremoniniR.1992. The chromosomes of Vicia faba: banding patterns and in situ hybridizations. Biologisches Zentralblatt 111: 188–203.

