Discrete models of autocrine cell communication in epithelial layers
- PMID: 12770871
- PMCID: PMC1302947
- DOI: 10.1016/S0006-3495(03)75093-0
Discrete models of autocrine cell communication in epithelial layers
Abstract
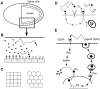
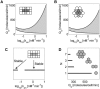
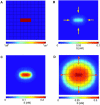
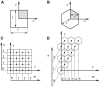
Pattern formation in epithelial layers heavily relies on cell communication by secreted ligands. Whereas the experimentally observed signaling patterns can be visualized at single-cell resolution, a biophysical framework for their interpretation is currently lacking. To this end, we develop a family of discrete models of cell communication in epithelial layers. The models are based on the introduction of cell-to-cell coupling coefficients that characterize the spatial range of intercellular signaling by diffusing ligands. We derive the coupling coefficients as functions of geometric, cellular, and molecular parameters of the ligand transport problem. Using these coupling coefficients, we analyze a nonlinear model of positive feedback between ligand release and binding. In particular, we study criteria of existence of the patterns consisting of clusters of a few signaling cells, as well as the onset of signal propagation. We use our model to interpret recent experimental studies of the EGFR/Rhomboid/Spitz module in Drosophila development.
Figures


Similar articles
-
Long-range signal transmission in autocrine relays.Biophys J. 2003 Feb;84(2 Pt 1):883-96. doi: 10.1016/S0006-3495(03)74906-6. Biophys J. 2003. PMID: 12547771 Free PMC article.
-
Autocrine, paracrine and juxtacrine signaling by EGFR ligands.Cell Signal. 2005 Oct;17(10):1183-93. doi: 10.1016/j.cellsig.2005.03.026. Cell Signal. 2005. PMID: 15982853 Review.
-
Transitions in the model of epithelial patterning.Dev Dyn. 2003 Jan;226(1):155-9. doi: 10.1002/dvdy.10218. Dev Dyn. 2003. PMID: 12508238
-
Autocrine signal transmission with extracellular ligand degradation.Phys Biol. 2009 Feb 20;6(1):016006. doi: 10.1088/1478-3975/6/1/016006. Phys Biol. 2009. PMID: 19234361
-
Morphogen propagation and action: towards molecular models.Semin Cell Dev Biol. 1999 Jun;10(3):297-302. doi: 10.1006/scdb.1999.0296. Semin Cell Dev Biol. 1999. PMID: 10441543 Review.
Cited by
-
Paracrine communication maximizes cellular response fidelity in wound signaling.Elife. 2015 Oct 8;4:e09652. doi: 10.7554/eLife.09652. Elife. 2015. PMID: 26448485 Free PMC article.
-
Compensated optimal grids for elliptic boundary-value problems.J Comput Phys. 2008 Oct 1;227(19):8622-8635. doi: 10.1016/j.jcp.2008.06.026. J Comput Phys. 2008. PMID: 19802366 Free PMC article.
-
Quantifying the Gurken morphogen gradient in Drosophila oogenesis.Dev Cell. 2006 Aug;11(2):263-72. doi: 10.1016/j.devcel.2006.07.004. Dev Cell. 2006. PMID: 16890165 Free PMC article.
-
Structure of the EGF receptor transactivation circuit integrates multiple signals with cell context.Mol Biosyst. 2010 Jul;6(7):1293-306. doi: 10.1039/c003921g. Epub 2010 May 10. Mol Biosyst. 2010. PMID: 20458382 Free PMC article.
-
Emergent patterns of growth controlled by multicellular form and mechanics.Proc Natl Acad Sci U S A. 2005 Aug 16;102(33):11594-9. doi: 10.1073/pnas.0502575102. Epub 2005 Jul 27. Proc Natl Acad Sci U S A. 2005. PMID: 16049098 Free PMC article.
References
-
- Amiri, A., and D. Stein. 2002. Dorsoventral patterning: a direct route from ovary to embryo. Curr. Biol. 12:R532–R534. - PubMed
-
- Bergmann, A., M. Tugentman, B. Shilo, and H. Steller. 2002. Regulation of cell number by MAPK-dependent control of apoptosis. a mechanism for trophic survival signaling. Dev. Cell. 2:159–170. - PubMed
-
- Bolouri, H., and E. H. Davidson. 2002. Modeling transcriptional regulatory networks. Bioessays. 24:1118–1129. - PubMed
-
- Brand, A. H., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 118:401–415. - PubMed
-
- Cahn, J. W., J. Mallet-Paret, and E. S. Van Vleck. 1998. Traveling wave solutions for systems of ODEs on a two-dimensional spatial lattice. SIAM J. Appl. Math. 59:455–493.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

