The flexibility of DNA double crossover molecules
- PMID: 12770888
- PMCID: PMC1302964
- DOI: 10.1016/S0006-3495(03)75110-8
The flexibility of DNA double crossover molecules
Abstract
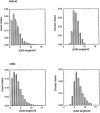
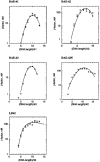
Double crossover molecules are DNA structures containing two Holliday junctions connected by two double helical arms. There are several types of double crossover molecules, differentiated by the relative orientations of their helix axes, parallel or antiparallel, and by the number of double helical half-turns (even or odd) between the two crossovers. They are found as intermediates in meiosis and they have been used extensively in structural DNA nanotechnology for the construction of one-dimensional and two-dimensional arrays and in a DNA nanomechanical device. Whereas the parallel double helical molecules are usually not well behaved, we have focused on the antiparallel molecules; antiparallel molecules with an even number of half-turns between crossovers (termed DAE molecules) produce a reporter strand when ligated, facilitating their characterization in a ligation cyclization assay. Hence, we have estimated the flexibility of antiparallel DNA double crossover molecules by means of ligation-closure experiments. We are able to show that these molecules are approximately twice as rigid as linear duplex DNA.
Figures







References
-
- Caruthers, M. H. 1985. Gene synthesis machines: DNA chemistry and its uses. Science. 230:281–285. - PubMed
-
- Fu, T.-J., and N. C. Seeman. 1993. DNA double crossover molecules. Biochemistry. 32:3211–3220. - PubMed
-
- Fu, T.-J., Y.-C. Tse-Dinh, and N. C. Seeman. 1994a. Holliday junction crossover topology. J. Mol. Biol. 236:91–105. - PubMed
-
- Fu, T.-J., B. Kemper, and N. C. Seeman. 1994b. Endonuclease VII cleavage of DNA double crossover molecules. Biochemistry. 33:3896–3905. - PubMed
-
- Hagerman, P. J. 1988. Flexibility of DNA. Annu. Rev. Biophys. Biophys. Chem. 17:265–286. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

