Molecular dynamics simulations of lignin peroxidase in solution
- PMID: 12770894
- PMCID: PMC1302970
- DOI: 10.1016/s0006-3495(03)75116-9
Molecular dynamics simulations of lignin peroxidase in solution
Abstract
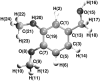
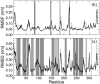
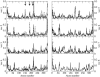
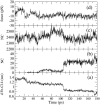
The dynamical and structural properties of lignin peroxidase and its Trp171Ala mutant have been investigated in aqueous solution using molecular dynamics (MD) simulations. In both cases, the enzyme retained its overall backbone structure and all its noncovalent interactions in the course of the MD simulations. Very interestingly, the analysis of the MD trajectories showed the presence of large fluctuations in correspondence of the residues forming the heme access channel; these movements enlarge the opening and facilitate the access of substrates to the enzyme active site. Moreover, steered molecular dynamics docking simulations have shown that lignin peroxidase natural substrate (veratryl alcohol) can easily approach the heme edge through the access channel.
Figures




Similar articles
-
The crystal structure of lignin peroxidase at 1.70 A resolution reveals a hydroxy group on the cbeta of tryptophan 171: a novel radical site formed during the redox cycle.J Mol Biol. 1999 Feb 26;286(3):809-27. doi: 10.1006/jmbi.1998.2507. J Mol Biol. 1999. PMID: 10024453
-
Addition of veratryl alcohol oxidase activity to manganese peroxidase by site-directed mutagenesis.Biochem Biophys Res Commun. 1999 Mar 24;256(3):500-4. doi: 10.1006/bbrc.1999.0360. Biochem Biophys Res Commun. 1999. PMID: 10080927
-
Lignin peroxidase ligand access channel dysfunction in the presence of atrazine.Sci Rep. 2018 Apr 16;8(1):5989. doi: 10.1038/s41598-018-24478-w. Sci Rep. 2018. PMID: 29662099 Free PMC article.
-
Manganese peroxidase.Met Ions Biol Syst. 2000;37:559-86. Met Ions Biol Syst. 2000. PMID: 10693145 Review. No abstract available.
-
Heme to protein linkages in mammalian peroxidases: impact on spectroscopic, redox and catalytic properties.Nat Prod Rep. 2007 Jun;24(3):571-84. doi: 10.1039/b604178g. Epub 2007 Mar 21. Nat Prod Rep. 2007. PMID: 17534531 Review. No abstract available.
Cited by
-
White Rot Fungi as Tools for the Bioremediation of Xenobiotics: A Review.J Fungi (Basel). 2024 Feb 21;10(3):167. doi: 10.3390/jof10030167. J Fungi (Basel). 2024. PMID: 38535176 Free PMC article. Review.
-
An overview of biomass conversion: exploring new opportunities.PeerJ. 2020 Jul 22;8:e9586. doi: 10.7717/peerj.9586. eCollection 2020. PeerJ. 2020. PMID: 32765969 Free PMC article.
-
Saprotrophic capabilities as functional traits to study functional diversity and resilience of ectomycorrhizal community.Oecologia. 2009 Oct;161(4):661-4. doi: 10.1007/s00442-009-1434-6. Epub 2009 Aug 15. Oecologia. 2009. PMID: 19685248
-
In silico exploration of lignin peroxidase for unraveling the degradation mechanism employing lignin model compounds.RSC Adv. 2021 Apr 20;11(24):14632-14653. doi: 10.1039/d0ra10840e. eCollection 2021 Apr 15. RSC Adv. 2021. PMID: 35423962 Free PMC article.
-
A molecular dynamics study of the ligand release path in yeast cytosine deaminase.Biophys J. 2007 Apr 1;92(7):2301-10. doi: 10.1529/biophysj.106.098921. Epub 2007 Jan 11. Biophys J. 2007. PMID: 17218460 Free PMC article.
References
-
- Allen, M. P., and D. J. Tildesly. 1989. Computer simulation of liquids. Oxford University Press, Oxford.
-
- Amadei, A., G. Chillemi, M. A. Ceruso, A. Grottesi, and A. Di Nola. 2000. Molecular dynamics simulations with constrained roto-translational motions: theoretical basis and statistical mechanical consistency. J. Chem. Phys. 112:9–23.
-
- Amadei, A., A. B. M. Linssen, and H. J. C. Berendsen. 1993. Essential dynamics of proteins. Prot. Struct. Funct. Gen. 17:412–425. - PubMed
-
- Andrawis, A., K. A. Johnson, and M. Tien. 1988. Studies on compound I formation of the lignin peroxidase from Phanerochaete chrysosporium. J. Biol. Chem. 263:1195–1198. - PubMed
-
- Baciocchi, E., M. F. Gerini, P. J. Harvey, O. Lanzalunga, and S. Mancinelli. 2000. Oxidation of aromatic sulfides by lignin peroxidase from Phanerochaete chrysosporium. Eur. J. Biochem. 267:2705–2710. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

