Functional studies and distribution define a family of transmembrane AMPA receptor regulatory proteins
- PMID: 12771129
- PMCID: PMC2199354
- DOI: 10.1083/jcb.200212116
Functional studies and distribution define a family of transmembrane AMPA receptor regulatory proteins
Abstract
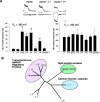
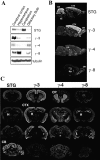
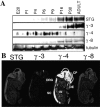
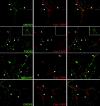
Functional expression of alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors in cerebellar granule cells requires stargazin, a member of a large family of four-pass transmembrane proteins. Here, we define a family of transmembrane AMPA receptor regulatory proteins (TARPs), which comprise stargazin, gamma-3, gamma-4, and gamma-8, but not related proteins, that mediate surface expression of AMPA receptors. TARPs exhibit discrete and complementary patterns of expression in both neurons and glia in the developing and mature central nervous system. In brain regions that express multiple isoforms, such as cerebral cortex, TARP-AMPA receptor complexes are strictly segregated, suggesting distinct roles for TARP isoforms. TARPs interact with AMPA receptors at the postsynaptic density, and surface expression of mature AMPA receptors requires a TARP. These studies indicate a general role for TARPs in controlling synaptic AMPA receptors throughout the central nervous system.
Figures

References
-
- Burgess, D.L., L.A. Gefrides, P.J. Foreman, and J.L. Noebels. 2001. A cluster of three novel Ca2+ channel gamma subunit genes on chromosome 19q13.4: evolution and expression profile of the gamma subunit gene family. Genomics. 71:339–350. - PubMed
-
- Chen, L., D.M. Chetkovich, R. Petralia, N. Sweeney, Y. Kawaski, R. Wenthold, D.S. Bredt, and R.A. Nicoll. 2000. Stargazin mediates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature. 408:936–943. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

