Overexpression of FABP7 in Down syndrome fetal brains is associated with PKNOX1 gene-dosage imbalance
- PMID: 12771203
- PMCID: PMC156729
- DOI: 10.1093/nar/gkg396
Overexpression of FABP7 in Down syndrome fetal brains is associated with PKNOX1 gene-dosage imbalance
Abstract
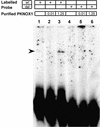
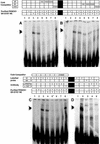
Suppression subtractive hybridization performed on Down syndrome (DS) fetal brains revealed a differentially expressed gene, FABP7, mapped to 6q22-23. FABP7 overexpression in DS brains was verified by real-time PCR (1.63-fold). To elucidate the molecular basis of FABP7 overexpression and establish the relationship with chromosome 21 trisomy, the FABP7 promoter was cloned by genomic inverse PCR. Comparison to the mouse ortholog revealed conservation of reported regulatory elements, among them a Pbx/POU binding site, known to be the target of PBX heteromeric complexes. PBX partners include homeobox-containing proteins, such as PKNOX1 (PREP1), a transcription factor mapping at 21q22.3. We report here: (i) overexpression of PKNOX1 in DS fetal brains; (ii) in vitro specific binding of PKNOX1 to the Pbx/POU site of the FABP7 promoter; (iii) in vivo FABP7 promoter trans-activation in cultured neuroblastoma cells caused by PKNOX1 overexpression. To our knowledge this is the first report of a direct relation between dosage imbalance of a chromosome 21 gene and altered expression of a downstream gene mapping on another chromosome. Given the role of FABP7 in the establishment, development and maintenance of the CNS, we suggest that the overexpression of FABP7 could contribute to DS-associated neurological disorders.
Figures


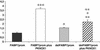
References
-
- Hassold T. and Jacobs,P. (1984) Trisomy in man. Annu. Rev. Genet., 18, 69–97. - PubMed
-
- Epstein C.J. (1995) Epilogue: toward twenty-first century with Down syndrome—a personal view of how far we have come and how far we can reasonably expect to go. Prog. Clin. Biol. Res., 393, 241–246. - PubMed
-
- Epstein C.J. (1986) The Consequences of Chromosome Imbalance: Principles, Mechanisms and Models. Cambridge University Press, New York.
-
- Hattori M., Fujiyama,A., Taylor,T.D., Watanabe,H., Yada,T., Parks,H.S., Toyoda,A., Ishii,K., Totoki,Y., Choi,D.K. et al. (2000) The DNA sequence of human chromosome 21. Nature, 405, 311–319. - PubMed
-
- Reymond A., Marigo,V., Yaylaoglu,M.B., Leoni. A., Ucla,C., Scamuffa,N., Cacciopoli,C., Dermitzakis,E.T., Lyle,R., Banfi,S. et al. (2002) Human chromosome 21 gene expression atlas in the mouse. Nature, 420, 582–586. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical
Research Materials

