Purification and characterisation of a novel DNA methyltransferase, M.AhdI
- PMID: 12771207
- PMCID: PMC156732
- DOI: 10.1093/nar/gkg399
Purification and characterisation of a novel DNA methyltransferase, M.AhdI
Abstract
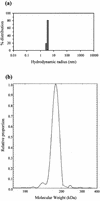
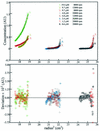
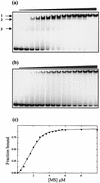
We have cloned the M and S genes of the restriction-modification (R-M) system AhdI and have purified the resulting methyltransferase to homogeneity. M.AhdI is found to form a 170 kDa tetrameric enzyme having a subunit stoichiometry M2S2 (where the M and S subunits are responsible for methylation and DNA sequence specificity, respectively). Sedimentation equilibrium experiments show that the tetrameric enzyme dissociates to form a heterodimer at low concentration, with K(d) approximately 2 microM. The intact (tetrameric) enzyme binds specifically to a 30 bp DNA duplex containing the AhdI recognition sequence GACN5GTC with high affinity (K(d) approximately 50 nM), but at low enzyme concentration the DNA binding activity is governed by the dissociation of the tetramer into dimers, leading to a sigmoidal DNA binding curve. In contrast, only non-specific binding is observed if the duplex lacks the recognition sequence. Methylation activity of the purified enzyme was assessed by its ability to prevent restriction by the cognate endonuclease. The subunit structure of the M.AhdI methyltransferase resembles that of type I MTases, in contrast to the R.AhdI endonuclease which is typical of type II systems. AhdI appears to be a novel R-M system with properties intermediate between simple type II systems and more complex type I systems, and may represent an intermediate in the evolution of R-M systems.
Figures

Similar articles
-
Shape and subunit organisation of the DNA methyltransferase M.AhdI by small-angle neutron scattering.J Mol Biol. 2007 May 25;369(1):177-185. doi: 10.1016/j.jmb.2007.03.012. Epub 2007 Mar 14. J Mol Biol. 2007. PMID: 17418232 Free PMC article.
-
A novel strategy for the expression and purification of the DNA methyltransferase, M.AhdI.Protein Expr Purif. 2004 Sep;37(1):236-42. doi: 10.1016/j.pep.2004.06.008. Protein Expr Purif. 2004. PMID: 15294304
-
DNA footprinting and biophysical characterization of the controller protein C.AhdI suggests the basis of a genetic switch.Nucleic Acids Res. 2004 Dec 8;32(21):6445-53. doi: 10.1093/nar/gkh975. Print 2004. Nucleic Acids Res. 2004. PMID: 15590905 Free PMC article.
-
Specificity of restriction endonucleases and DNA modification methyltransferases a review (Edition 3).Gene. 1990 Aug 16;92(1-2):1-248. doi: 10.1016/0378-1119(90)90486-b. Gene. 1990. PMID: 2172084 Review.
-
A symmetrical model for the domain structure of type I DNA methyltransferases.J Mol Biol. 1994 Oct 14;243(1):1-5. doi: 10.1006/jmbi.1994.1624. J Mol Biol. 1994. PMID: 7932730 Review.
Cited by
-
Transcription regulation of the type II restriction-modification system AhdI.Nucleic Acids Res. 2008 Mar;36(5):1429-42. doi: 10.1093/nar/gkm1116. Epub 2008 Jan 18. Nucleic Acids Res. 2008. PMID: 18203750 Free PMC article.
-
Type II restriction endonucleases--a historical perspective and more.Nucleic Acids Res. 2014 Jul;42(12):7489-527. doi: 10.1093/nar/gku447. Epub 2014 May 30. Nucleic Acids Res. 2014. PMID: 24878924 Free PMC article. Review.
-
MmeI: a minimal Type II restriction-modification system that only modifies one DNA strand for host protection.Nucleic Acids Res. 2008 Nov;36(20):6558-70. doi: 10.1093/nar/gkn711. Epub 2008 Oct 17. Nucleic Acids Res. 2008. PMID: 18931376 Free PMC article.
-
Highlights of the DNA cutters: a short history of the restriction enzymes.Nucleic Acids Res. 2014 Jan;42(1):3-19. doi: 10.1093/nar/gkt990. Epub 2013 Oct 18. Nucleic Acids Res. 2014. PMID: 24141096 Free PMC article.
-
Type I restriction enzymes and their relatives.Nucleic Acids Res. 2014 Jan;42(1):20-44. doi: 10.1093/nar/gkt847. Epub 2013 Sep 24. Nucleic Acids Res. 2014. PMID: 24068554 Free PMC article. Review.
References
-
- Dryden D.T.F. (1999) Bacterial DNA methyltransferases. In Cheng,X. and Blumenthal,R.M. (eds), S-adenosyl Methionine Dependent Methyltransferases: Structure and Function. World Scientific Publishing, New York, NY, pp. 283–340.
-
- Hornby D.P. (1993) DNA methyltransferases. In Methods in Molecular Biology, Vol. 16: Enzymes of Molecular Biology. Humana Press, Totowa, NJ.
-
- Dryden D.T.F., Cooper,L.P. and Murray,N.E. (1993) Purification and characterization of the methyltransferase from the Type-1 restriction and modification system of E.coli K12. J. Biol. Chem., 268, 13228–13236. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

