Solution structure of the influenza A virus cRNA promoter: implications for differential recognition of viral promoter structures by RNA-dependent RNA polymerase
- PMID: 12771209
- PMCID: PMC156722
- DOI: 10.1093/nar/gkg387
Solution structure of the influenza A virus cRNA promoter: implications for differential recognition of viral promoter structures by RNA-dependent RNA polymerase
Abstract
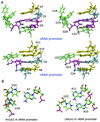
Influenza A virus replication requires the interaction of viral RNA-dependent RNA polymerase (RdRp) with promoters in both the RNA genome (vRNA) and the full-length complementary RNA (cRNA) which serve as templates for the generation of new vRNAs. Although RdRp binds both promoters effectively, it must also discriminate between them because they serve different functional roles in the viral life cycle. Even though the inherent asymmetry between two RNA promoters is considered as a cause of the differential recognition by the RdRp, the structural basis for the ability of the RdRp to recognize the RNA promoters and discriminate effectively between them remains unsolved. Here we report the structure of the cRNA promoter of influenza A virus as determined by heteronuclear magnetic resonance spectroscopy. The terminal region is extremely unstable and does not have a rigid structure. The major groove of the internal loop is widened by the displacement of a novel A*(UU) motif toward the minor groove. These internal loop residues show distinguishable dynamic characters, with differing motional timescales for each residue. Comparison of the cRNA promoter structure with that of the vRNA promoter reveals common structural and dynamic elements in the internal loop, but also differences that provide insight into how the viral RdRp differentially recognizes the cRNA and vRNA promoters.
Figures





Similar articles
-
Mutation of an Influenza Virus Polymerase 3' RNA Promoter Binding Site Inhibits Transcription Elongation.J Virol. 2020 Jun 16;94(13):e00498-20. doi: 10.1128/JVI.00498-20. Print 2020 Jun 16. J Virol. 2020. PMID: 32295915 Free PMC article.
-
The RNA polymerase of influenza a virus is stabilized by interaction with its viral RNA promoter.J Virol. 2002 Jul;76(14):7103-13. doi: 10.1128/jvi.76.14.7103-7113.2002. J Virol. 2002. PMID: 12072510 Free PMC article.
-
Differential activation of influenza A virus endonuclease activity is dependent on multiple sequence differences between the virion RNA and cRNA promoters.J Virol. 2002 Feb;76(4):2019-23. doi: 10.1128/jvi.76.4.2019-2023.2002. J Virol. 2002. PMID: 11799200 Free PMC article.
-
Structural insights into RNA synthesis by the influenza virus transcription-replication machine.Virus Res. 2017 Apr 15;234:103-117. doi: 10.1016/j.virusres.2017.01.013. Epub 2017 Jan 20. Virus Res. 2017. PMID: 28115197 Review.
-
Transcription and replication of the influenza a virus genome.Acta Virol. 2000 Oct;44(5):273-82. Acta Virol. 2000. PMID: 11252672 Review.
Cited by
-
Structural characterization of the viral and cRNA panhandle motifs from the infectious salmon anemia virus.J Virol. 2011 Dec;85(24):13398-408. doi: 10.1128/JVI.06250-11. Epub 2011 Oct 12. J Virol. 2011. PMID: 21994446 Free PMC article.
-
Structural and Functional Motifs in Influenza Virus RNAs.Front Microbiol. 2018 Mar 29;9:559. doi: 10.3389/fmicb.2018.00559. eCollection 2018. Front Microbiol. 2018. PMID: 29651275 Free PMC article. Review.
-
The N-terminal region of influenza virus polymerase PB1 adjacent to the PA binding site is involved in replication but not transcription of the viral genome.Front Microbiol. 2013 Dec 18;4:398. doi: 10.3389/fmicb.2013.00398. eCollection 2013. Front Microbiol. 2013. PMID: 24391632 Free PMC article.
-
Influenza virus segment 5 (+)RNA - secondary structure and new targets for antiviral strategies.Sci Rep. 2017 Nov 8;7(1):15041. doi: 10.1038/s41598-017-15317-5. Sci Rep. 2017. PMID: 29118447 Free PMC article.
-
Characterizing hydrogen bonds in intact RNA from MS2 bacteriophage using magic angle spinning NMR.Biophys Rep (N Y). 2021 Sep 29;1(2):100027. doi: 10.1016/j.bpr.2021.100027. eCollection 2021 Dec 8. Biophys Rep (N Y). 2021. PMID: 36425459 Free PMC article.
References
-
- Lamb R.A. and Choppin,P.W. (1983) The gene structure and replication of influenza virus. Annu. Rev. Biochem., 52, 467–506. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

