Optimisation of the 10-23 DNAzyme-substrate pairing interactions enhanced RNA cleavage activity at purine-cytosine target sites
- PMID: 12771215
- PMCID: PMC156713
- DOI: 10.1093/nar/gkg378
Optimisation of the 10-23 DNAzyme-substrate pairing interactions enhanced RNA cleavage activity at purine-cytosine target sites
Abstract
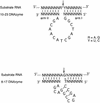
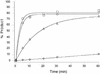
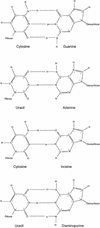
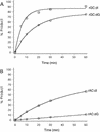
The 10-23 RNA cleaving DNAzyme has been shown to cleave any purine-pyrimidine (RY) junction under simulated physiological conditions. In this study, we systematically examine the DNAzymes relative activity against different RY combinations in order to determine the hierarchy of substrate core dinucleotide sequence susceptibility. The reactivity of each substrate dinucleotide compared in the same background sequence with the appropriately matched DNAzyme was found to follow the scheme AU = GU >> or = GC >> AC. The relatively poor activity of the DNAzyme against AC and GC containing substrates was found to be improved substantially by modifications to the binding domain which subtly weaken its interaction with the substrate core. The most effective modification resulting in rate enhancement of up to 200-fold, was achieved by substitution of deoxyguanine with deoxyinosine such that the base pair interaction with the RNA substrates core C is reduced from three hydrogen bonds to two. The increased cleavage activity generated by this modification could be important for application of the 10-23 DNAzyme particularly when the target site core is an AC dinucleotide.
Figures
Similar articles
-
The influence of arm length asymmetry and base substitution on the activity of the 10-23 DNA enzyme.Antisense Nucleic Acid Drug Dev. 2000 Oct;10(5):323-32. doi: 10.1089/oli.1.2000.10.323. Antisense Nucleic Acid Drug Dev. 2000. PMID: 11079572
-
Nucleic acid sequence analysis using DNAzymes.Methods Mol Biol. 2004;252:291-302. doi: 10.1385/1-59259-746-7:291. Methods Mol Biol. 2004. PMID: 15017058
-
A complex RNA-cleaving DNAzyme that can efficiently cleave a pyrimidine-pyrimidine junction.J Mol Biol. 2010 Jul 23;400(4):689-701. doi: 10.1016/j.jmb.2010.05.047. Epub 2010 May 26. J Mol Biol. 2010. PMID: 20630470
-
Molecular Features and Metal Ions That Influence 10-23 DNAzyme Activity.Molecules. 2020 Jul 7;25(13):3100. doi: 10.3390/molecules25133100. Molecules. 2020. PMID: 32646019 Free PMC article. Review.
-
A versatile endoribonuclease mimic made of DNA: characteristics and applications of the 8-17 RNA-cleaving DNAzyme.Chembiochem. 2010 May 3;11(7):866-79. doi: 10.1002/cbic.200900786. Chembiochem. 2010. PMID: 20213779 Review.
Cited by
-
Selective Characterization of mRNA 5' End-Capping by DNA Probe-Directed Enrichment with Site-Specific Endoribonucleases.ACS Pharmacol Transl Sci. 2023 Oct 18;6(11):1692-1702. doi: 10.1021/acsptsci.3c00157. eCollection 2023 Nov 10. ACS Pharmacol Transl Sci. 2023. PMID: 37974627 Free PMC article.
-
Locked nucleoside analogues expand the potential of DNAzymes to cleave structured RNA targets.BMC Mol Biol. 2006 Jun 5;7:19. doi: 10.1186/1471-2199-7-19. BMC Mol Biol. 2006. PMID: 16753066 Free PMC article.
-
Advances and Trends in miRNA Analysis Using DNAzyme-Based Biosensors.Biosensors (Basel). 2023 Aug 29;13(9):856. doi: 10.3390/bios13090856. Biosensors (Basel). 2023. PMID: 37754090 Free PMC article. Review.
-
Primer fabrication using polymerase mediated oligonucleotide synthesis.BMC Genomics. 2009 Jul 31;10:344. doi: 10.1186/1471-2164-10-344. BMC Genomics. 2009. PMID: 19643029 Free PMC article.
-
Influence of LNA modifications on the activity of the 10-23 DNAzyme.RSC Adv. 2025 Apr 23;15(17):13031-13040. doi: 10.1039/d5ra00161g. eCollection 2025 Apr 22. RSC Adv. 2025. PMID: 40271416 Free PMC article.
References
-
- Santoro S.W. and Joyce,G.F. (1998) Mechanism and utility of an RNA-cleaving DNA enzyme. Biochemistry, 37, 13330–13342. - PubMed
-
- Sun L.Q., Cairns,M.J., Gerlach,W., Witherington,C., Wang,L. and King,A. (1999) Suppression of smooth muscle cell proliferation by a c-myc RNA-cleaving deoxyribozyme. J. Biol. Chem., 274, 17236–17241. - PubMed
-
- Santiago F.S., Kavurma,M.M., Lowe,H.C., Chesterman,C.N., Baker,A., Atkins,D.G. and Khachigian,L.M. (1999) New DNA enzyme targeting Egr-1 mRNA inhibits vascular smooth muscle proliferation and regrowth after injury. Nature Med., 5, 1264–1269. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

