Structural basis for SH3 domain-mediated high-affinity binding between Mona/Gads and SLP-76
- PMID: 12773374
- PMCID: PMC156755
- DOI: 10.1093/emboj/cdg258
Structural basis for SH3 domain-mediated high-affinity binding between Mona/Gads and SLP-76
Abstract
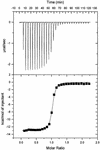
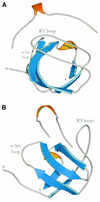
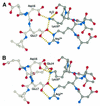
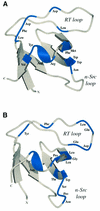
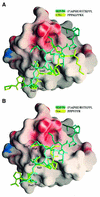
SH3 domains are protein recognition modules within many adaptors and enzymes. With more than 500 SH3 domains in the human genome, binding selectivity is a key issue in understanding the molecular basis of SH3 domain interactions. The Grb2-like adaptor protein Mona/Gads associates stably with the T-cell receptor signal transducer SLP-76. The crystal structure of a complex between the C-terminal SH3 domain (SH3C) of Mona/Gads and a SLP-76 peptide has now been solved to 1.7 A. The peptide lacks the canonical SH3 domain binding motif P-x-x-P and does not form a frequently observed poly-proline type II helix. Instead, it adopts a clamp-like shape around the circumfence of the SH3C beta-barrel. The central R-x-x-K motif of the peptide forms a 3(10) helix and inserts into a negatively charged double pocket on the SH3C while several other residues complement binding through hydrophobic interactions, creating a short linear SH3C binding epitope of uniquely high affinity. Interestingly, the SH3C displays ion-dependent dimerization in the crystal and in solution, suggesting a novel mechanism for the regulation of SH3 domain functions.
Figures





References
-
- Berry D.M., Nash,P., Liu,S.K., Pawson,T. and McGlade,C.J. (2002) A high-affinity Arg–X–X–Lys SH3 binding motif confers specificity for the interaction between Gads and SLP-76 in T cell signaling. Curr. Biol., 12, 1336–1341. - PubMed
-
- Bork P., Schultz,J. and Ponting,C.P. (1997) Cytoplasmic signalling domains: the next generation. Trends Biochem. Sci., 22, 296–298. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

