Crystal structure of tRNA(m1G37)methyltransferase: insights into tRNA recognition
- PMID: 12773376
- PMCID: PMC156765
- DOI: 10.1093/emboj/cdg269
Crystal structure of tRNA(m1G37)methyltransferase: insights into tRNA recognition
Abstract
tRNA(m(1)G37)methyltransferase (TrmD) catalyzes the transfer of a methyl group from S-adenosyl-L- methionine (AdoMet) to G(37) within a subset of bacterial tRNA species, which have a G residue at the 36th position. The modified guanosine is adjacent to and 3' of the anticodon and is essential for the maintenance of the correct reading frame during translation. Here we report four crystal structures of TrmD from Haemophilus influenzae, as binary complexes with either AdoMet or S-adenosyl-L-homocysteine (AdoHcy), as a ternary complex with AdoHcy and phosphate, and as an apo form. This first structure of TrmD indicates that it functions as a dimer. It also suggests the binding mode of G(36)G(37) in the active site of TrmD and the catalytic mechanism. The N-terminal domain has a trefoil knot, in which AdoMet or AdoHcy is bound in a novel, bent conformation. The C-terminal domain shows structural similarity to trp repressor. We propose a plausible model for the TrmD(2)-tRNA(2) complex, which provides insights into recognition of the general tRNA structure by TrmD.
Figures
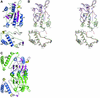
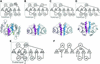



References
-
- Anantharaman V, Koonin,E.V. and Aravind,L. (2002) SPOUT: a class of methyltransferases that includes spoU and trmD RNA methylase superfamilies and novel superfamilies of predicted prokaryotic RNA methylases. J. Mol. Microbiol. Biotechnol., 4, 71–75. - PubMed
-
- Batey R.T., Rambo,R.P., Lucast,L., Rha,B. and Doudna,J.A. (2000) Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science, 287, 1232–1239. - PubMed
-
- Battiste J.L., Mao,H., Rao,N.S., Tan,R., Muhandiram,D.R., Kay,L.E., Frankel,A.D. and Williamson,J.R. (1996) α helix–RNA major groove recognition in an HIV-1 Rev peptide–RRE RNA complex. Science, 273, 1547–1551. - PubMed
-
- Björk G.R. (1998) Modified nucleosides in positions 34 and 37 of tRNAs and their predicted coding capacities. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 577–581.
-
- Björk G.R., Wikström,P.M. and Byström,A.S. (1989) Prevention of translational frameshifting by the modified nucleoside 1-methyl guanosine. Science, 244, 986–989. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

