Rabaptin-5alpha/rabaptin-4 serves as a linker between rab4 and gamma(1)-adaptin in membrane recycling from endosomes
- PMID: 12773381
- PMCID: PMC156754
- DOI: 10.1093/emboj/cdg257
Rabaptin-5alpha/rabaptin-4 serves as a linker between rab4 and gamma(1)-adaptin in membrane recycling from endosomes
Abstract
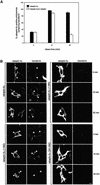
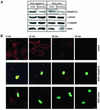
Rab4 regulates recycling from early endosomes. We investigated the role of the rab4 effector rabaptin-5alpha and its putative partner gamma(1)-adaptin in membrane recycling. We found that rabaptin-5alpha forms a ternary complex with the gamma(1)-sigma(1) subcomplex of AP-1, via a direct interaction with the gamma(1)-subunit. The binding site for gamma(1)-adaptin is in the hinge region of rabaptin-5alpha, which is distinct from rab4- and rab5-binding domains. Endogenous or ectopically expressed gamma(1)- adaptin localized to both the trans-Golgi network and endosomes. Co-expressed rabaptin-5alpha and gamma(1)-adaptin, however, co-localized in a rab4-dependent manner on recycling endosomes. Transfection of rabaptin-5alpha caused enlarged endosomes and delayed recycling of transferrin. RNAi of rab4 had an opposing effect on transferrin recycling. Collectively, our data show that rab4-GTP acts as a scaffold for a rabaptin-5alpha- gamma(1)-adaptin complex on recycling endosomes and that interactions between rab4, rabaptin-5alpha and gamma(1)-adaptin regulate membrane recycling.
Figures

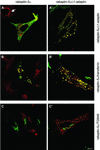
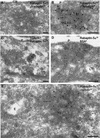
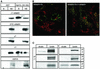

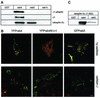
References
-
- Brummelkamp T.R., Bernards,R. and Agami,R. (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science, 296, 550–553. - PubMed
-
- Bucci C., Parton,R., Mather,I., Stunnenberg,H., Simons,K. and Zerial,M. (1992) The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell, 70, 715–728. - PubMed
-
- Collins B.M., McCoy,A.J., Kent,H.M., Evans,P.R. and Owen,D.J. (2002) Molecular architecture and functional model of the endocytic AP2 complex. Cell, 109, 523–535. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

