Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates
- PMID: 12773398
- PMCID: PMC156746
- DOI: 10.1093/emboj/cdg249
Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates
Abstract
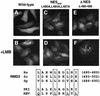
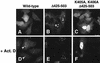
60S and 40S ribosomal subunits are assembled in the nucleolus and exported from the nucleus to the cytoplasm independently of each other. We show that in vertebrate cells, transport of both subunits requires the export receptor CRM1 and Ran.GTP. Export of 60S subunits is coupled with that of the nucleo- cytoplasmic shuttling protein NMD3. Human NMD3 (hNMD3) contains a CRM-1-dependent leucine-rich nuclear export signal (NES) and a complex, dispersed nuclear localization signal (NLS), the basic region of which is also required for nucleolar accumulation. When present in Xenopus oocytes, both wild-type and export-defective mutant hNMD3 proteins bind to newly made nuclear 60S pre-export particles at a late step of subunit maturation. The export-defective hNMD3, but not the wild-type protein, inhibits export of 60S subunits from oocyte nuclei. These results indicate that the NES mutant protein competes with endogenous wild-type frog NMD3 for binding to nascent 60S subunits, thereby preventing their export. We propose that NMD3 acts as an adaptor for CRM1-Ran.GTP-mediated 60S subunit export, by a mechanism that is conserved from vertebrates to yeast.
Figures





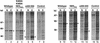



Similar articles
-
Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway.J Cell Sci. 2003 Jun 15;116(Pt 12):2409-19. doi: 10.1242/jcs.00464. Epub 2003 Apr 30. J Cell Sci. 2003. PMID: 12724356
-
Novel interaction of the 60S ribosomal subunit export adapter Nmd3 at the nuclear pore complex.J Biol Chem. 2007 May 11;282(19):14028-37. doi: 10.1074/jbc.M700256200. Epub 2007 Mar 8. J Biol Chem. 2007. PMID: 17347149
-
Nuclear export of ribosomal subunits.Trends Biochem Sci. 2002 Nov;27(11):580-5. doi: 10.1016/s0968-0004(02)02208-9. Trends Biochem Sci. 2002. PMID: 12417134 Review.
-
Reengineering ribosome export.Mol Biol Cell. 2009 Mar;20(5):1545-54. doi: 10.1091/mbc.e08-10-1000. Epub 2009 Jan 14. Mol Biol Cell. 2009. PMID: 19144820 Free PMC article.
-
Nuclear export and cytoplasmic maturation of ribosomal subunits.FEBS Lett. 2007 Jun 19;581(15):2783-93. doi: 10.1016/j.febslet.2007.05.013. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17509569 Review.
Cited by
-
How SARS-CoV-2 and Other Viruses Build an Invasion Route to Hijack the Host Nucleocytoplasmic Trafficking System.Cells. 2021 Jun 7;10(6):1424. doi: 10.3390/cells10061424. Cells. 2021. PMID: 34200500 Free PMC article. Review.
-
Mutational Analysis of the Nsa2 N-Terminus Reveals Its Essential Role in Ribosomal 60S Subunit Assembly.Int J Mol Sci. 2020 Nov 30;21(23):9108. doi: 10.3390/ijms21239108. Int J Mol Sci. 2020. PMID: 33266193 Free PMC article.
-
GNL3L Is a Nucleo-Cytoplasmic Shuttling Protein: Role in Cell Cycle Regulation.PLoS One. 2015 Aug 14;10(8):e0135845. doi: 10.1371/journal.pone.0135845. eCollection 2015. PLoS One. 2015. PMID: 26274615 Free PMC article.
-
A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export.PLoS Biol. 2010 Oct 26;8(10):e1000522. doi: 10.1371/journal.pbio.1000522. PLoS Biol. 2010. PMID: 21048991 Free PMC article.
-
Nuclear Export of Pre-Ribosomal Subunits Requires Dbp5, but Not as an RNA-Helicase as for mRNA Export.PLoS One. 2016 Feb 12;11(2):e0149571. doi: 10.1371/journal.pone.0149571. eCollection 2016. PLoS One. 2016. PMID: 26872259 Free PMC article.
References
-
- Allison L.A., Romaniuk,P.J. and Bakken,A.H. (1991) RNA–protein interactions of stored 5S RNA with TFIIIA and ribosomal protein L5 during Xenopus oogenesis. Dev. Biol., 144, 129–144. - PubMed
-
- Andersen J.S., Lyon,C.E., Fox,A.H., Leung,A.K., Lam,Y.W., Steen,H., Mann,M. and Lamond,A.I. (2002) Directed proteomic analysis of the human nucleolus. Curr. Biol., 12, 1–11. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

