The genome-linked protein VPg of the Norwalk virus binds eIF3, suggesting its role in translation initiation complex recruitment
- PMID: 12773399
- PMCID: PMC156748
- DOI: 10.1093/emboj/cdg251
The genome-linked protein VPg of the Norwalk virus binds eIF3, suggesting its role in translation initiation complex recruitment
Abstract
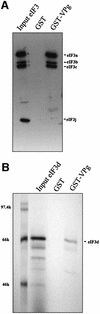
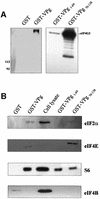
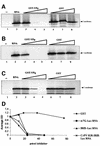
The positive-strand RNA genomes of caliciviruses are not capped, but are instead covalently linked at their 5' ends to a viral protein called VPg. The lack of a cap structure typical of eukaryotic mRNA and absence of an internal ribosomal entry site suggest that VPg may function in translation initiation on calicivirus RNA. This hypothesis was tested by analyzing binding of Norwalk virus VPg to translation initiation factors. The eIF3d subunit of eIF3 was identified as a binding partner of VPg by yeast two-hybrid analysis. VPg bound to purified mammalian eIF3 and to eIF3 in mammalian cell lysates. To test the effects of the VPg- eIF3 interaction on translation, VPg was added to cell-free translation reactions programmed with either capped reporter RNA, an RNA containing an EMCV internal ribosomal entry site (IRES) or an RNA with a cricket paralysis virus IRES. VPg inhibited translation of all reporter RNAs in a dose-dependent manner. Together, the data suggest that VPg may play a role in initiating translation on calicivirus RNA through unique protein-protein interactions with the translation machinery.
Figures

References
-
- Asano K., Kinzy,T.G., Merrick,W.C. and Hershey,J.W. (1997a) Conservation and diversity of eukaryotic translation initiation factor eIF3. J. Biol. Chem., 272, 1101–1109. - PubMed
-
- Asano K., Vornlocher,H.P., Richter-Cook,N.J., Merrick,W.C., Hinnebusch,A.G. and Hershey,J.W. (1997b) Structure of cDNAs encoding human eukaryotic initiation factor 3 subunits. Possible roles in RNA binding and macromolecular assembly. J. Biol. Chem., 272, 27042–27052. - PubMed
-
- Brown-Luedi M.L., Meyer,L.J., Milburn,S.C., Yau,P.M., Corbett,S. and Hershey,J.W. (1982) Protein synthesis initiation factors from human HeLa cells and rabbit reticulocytes are similar: comparison of protein structure, activities and immunochemical properties. Biochemistry, 21, 4202–4206. - PubMed
-
- Burroughs J.N. and Brown,F. (1978) Presence of a covalently linked protein on calicivirus RNA. J. Gen. Virol., 41, 443–446. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

