Cotranscriptional processing of Drosophila histone mRNAs
- PMID: 12773550
- PMCID: PMC156150
- DOI: 10.1128/MCB.23.12.4046-4055.2003
Cotranscriptional processing of Drosophila histone mRNAs
Abstract
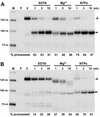
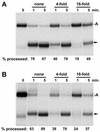
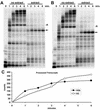
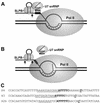
The 3' ends of metazoan histone mRNAs are generated by specialized processing machinery that cleaves downstream of a conserved stem-loop structure. To examine how this reaction might be influenced by transcription, we used a Drosophila melanogaster in vitro system that supports both processes. In this system the complete synthesis of histone mRNA, including transcription initiation and elongation, followed by 3' end formation, occurred at a physiologically significant rate. Processing of free transcripts was efficient and occurred with a t(1/2) of less than 1 min. Divalent cations were not required, but nucleoside triphosphates (NTPs) stimulated the rate of cleavage slightly. Isolated elongation complexes encountered a strong arrest site downstream of the mature histone H4 3' end. In the presence of NTPs, transcripts in these arrested complexes were processed at a rate similar to that of free RNA. Removal of NTPs dramatically reduced this rate, potentially due to concealment of the U7 snRNP binding element. The arrest site was found to be a conserved feature located 32 to 35 nucleotides downstream of the processing site on the H4, H2b, and H3 genes. The significance of the newly discovered arrest sites to our understanding of the coupling between transcription and RNA processing on the one hand and histone gene expression on the other is discussed.
Figures





References
-
- Bentley, D. 2002. The mRNA assembly line: transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 14:336-342. - PubMed
-
- Cramer, P., A. Srebrow, S. Kadener, S. Werbajh, M. de la Mata, G. Melen, G. Nogues, and A. R. Kornblihtt. 2001. Coordination between transcription and pre-mRNA processing. FEBS Lett. 498:179-182. - PubMed
-
- Dominski, Z., and W. F. Marzluff. 1999. Formation of the 3′ end of histone mRNA. Gene 239:1-14. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
