Role of the promoter in maintaining transcriptionally active chromatin structure and DNA methylation patterns in vivo
- PMID: 12773559
- PMCID: PMC156144
- DOI: 10.1128/MCB.23.12.4150-4161.2003
Role of the promoter in maintaining transcriptionally active chromatin structure and DNA methylation patterns in vivo
Abstract
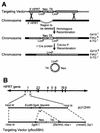
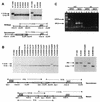
Establishment and maintenance of differential chromatin structure between transcriptionally competent and repressed genes are critical aspects of transcriptional regulation. The elements and mechanisms that mediate formation and maintenance of these chromatin states in vivo are not well understood. To examine the role of the promoter in maintaining chromatin structure and DNA methylation patterns of the transcriptionally active X-linked HPRT locus, 323 bp of the endogenous human HPRT promoter (from position -222 to +102 relative to the translation start site) was replaced by plasmid sequences by homologous recombination in cultured HT-1080 male fibrosarcoma cells. The targeted cells, which showed no detectable HPRT transcription, were then assayed for effects on DNase I hypersensitivity, general DNase I sensitivity, and DNA methylation patterns across the HPRT locus. In cells carrying the deletion, significantly diminished DNase I hypersensitivity in the 5' flanking region was observed compared to that in parental HT-1080 cells. However, general DNase I sensitivity and DNA methylation patterns were found to be very similar in the mutated cells and in the parental cells. These findings suggest that the promoter and active transcription play a relatively limited role in maintaining transcriptionally potentiated epigenetic states.
Figures

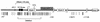



References
-
- Bender, M. A., M. Bulger, J. Close, and M. Groudine. 2000. Beta-globin gene switching and DNase I sensitivity of the endogenous beta-globin locus in mice do not require the locus control region. Mol. Cell 5:387-393. - PubMed
-
- Brandeis, M., D. Frank, I. Keshet, Z. Siegfried, M. Mendelsohn, A. Nemes, V. Temper, A. Razin, and H. Cedar. 1994. Sp1 elements protect a CpG island from de novo methylation. Nature 371:435-438. - PubMed
-
- Brown, C. J., A. Ballabio, J. L. Rupert, R. G. Lafreniere, M. Grompe, R. Tonlorenzi, and H. F. Willard. 1991. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349:38-44. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
