The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively
- PMID: 12773575
- PMCID: PMC156149
- DOI: 10.1128/MCB.23.12.4344-4355.2003
The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively
Abstract
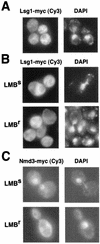
We characterized two essential putative GTPases, Nog1p and Lsg1p, that are found associated with free 60S ribosomal subunits affinity purified with the nuclear export adapter Nmd3p. Nog1p and Lsg1p are nucleolar and cytoplasmic, respectively, and are not simultaneously on the same particle, reflecting the path of Nmd3p shuttling in and out of the nucleus. Conditional mutants of both NOG1 and LSG1 are defective in 60S subunit biogenesis and display diminished levels of 60S subunits at restrictive temperature. Mutants of both genes also accumulate the 60S ribosomal reporter Rpl25-eGFP in the nucleolus, suggesting that both proteins are needed for subunit export from the nucleolus. Since Lsg1p is cytoplasmic, its role in nuclear export is likely to be indirect. We suggest that Lsg1p is needed to recycle an export factor(s) that shuttles from the nucleus associated with the nascent 60S subunit.
Figures








Similar articles
-
Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p.EMBO J. 2005 Feb 9;24(3):567-79. doi: 10.1038/sj.emboj.7600547. Epub 2005 Jan 20. EMBO J. 2005. PMID: 15660131 Free PMC article.
-
Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit.J Cell Biol. 2000 Nov 27;151(5):1057-66. doi: 10.1083/jcb.151.5.1057. J Cell Biol. 2000. PMID: 11086007 Free PMC article.
-
Nuclear recycling of the pre-60S ribosomal subunit-associated factor Arx1 depends on Rei1 in Saccharomyces cerevisiae.Mol Cell Biol. 2006 May;26(10):3718-27. doi: 10.1128/MCB.26.10.3718-3727.2006. Mol Cell Biol. 2006. PMID: 16648468 Free PMC article.
-
Nuclear export of ribosomal subunits.Trends Biochem Sci. 2002 Nov;27(11):580-5. doi: 10.1016/s0968-0004(02)02208-9. Trends Biochem Sci. 2002. PMID: 12417134 Review.
-
Ribosome biogenesis: stripping for AAAction?Curr Biol. 2001 Sep 4;11(17):R710-2. doi: 10.1016/s0960-9822(01)00416-x. Curr Biol. 2001. PMID: 11553347 Review.
Cited by
-
Saccharomyces cerevisiae Rbg1 protein and its binding partner Gir2 interact on Polyribosomes with Gcn1.Eukaryot Cell. 2009 Jul;8(7):1061-71. doi: 10.1128/EC.00356-08. Epub 2009 May 15. Eukaryot Cell. 2009. PMID: 19448108 Free PMC article.
-
The assembly factor Erb1 functions in multiple remodeling events during 60S ribosomal subunit assembly in S. cerevisiae.Nucleic Acids Res. 2017 May 5;45(8):4853-4865. doi: 10.1093/nar/gkw1361. Nucleic Acids Res. 2017. PMID: 28115637 Free PMC article.
-
Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S preribosomes.Mol Cell Biol. 2005 Dec;25(23):10419-32. doi: 10.1128/MCB.25.23.10419-10432.2005. Mol Cell Biol. 2005. PMID: 16287855 Free PMC article.
-
A functional network involved in the recycling of nucleocytoplasmic pre-60S factors.J Cell Biol. 2006 May 8;173(3):349-60. doi: 10.1083/jcb.200510080. Epub 2006 May 1. J Cell Biol. 2006. PMID: 16651379 Free PMC article.
-
The essential WD-repeat protein Rsa4p is required for rRNA processing and intra-nuclear transport of 60S ribosomal subunits.Nucleic Acids Res. 2005 Oct 12;33(18):5728-39. doi: 10.1093/nar/gki887. Print 2005. Nucleic Acids Res. 2005. PMID: 16221974 Free PMC article.
References
-
- Bassler, J., P. Grandi, O. Gadal, T. Lessmann, E. Petfalski, D. Tollervey, J. Lechner, and E. Hurt. 2001. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell 8:517-529. - PubMed
-
- Bécam, A. M., F. Nasr, W. J. Racki, M. Zagulski, and C. J. Herbert. 2001. Ria1p (Ynl163c), a protein similar to elongation factors 2, is involved in the biogenesis of the 60S subunit of the ribosome in Saccharomyces cerevisiae. Mol. Gen. Genet. 266:454-462. - PubMed
-
- Dragon, F., J. E. G. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417:967-970. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
