Getting a grip on non-native proteins
- PMID: 12776175
- PMCID: PMC1319208
- DOI: 10.1038/sj.embor.embor869
Getting a grip on non-native proteins
Abstract
It is an underappreciated fact that non-native polypeptides are prevalent in the cellular environment. Native proteins have the folded structure, assembled state and cellular localization required for activity. By contrast, non-native proteins lack function and are particularly prone to aggregation because hydrophobic residues that are normally buried are exposed on their surfaces. These unstable entities include polypeptides that are undergoing synthesis, transport to and translocation across membranes, and those that are unfolded before degradation. Non-native proteins are normal, biologically relevant components of a healthy cell, except in cases in which their misfolding results from disease-causing mutations or adverse extrinsic factors. Here, we explore the nature and occurrence of non-native proteins, and describe the diverse families of molecular chaperones and coordinated cellular responses that have evolved to prevent their misfolding and aggregation, thereby maintaining quality control over these potentially damaging protein species.
Figures
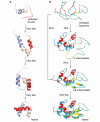

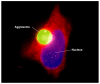



References
-
- Chernoff Y.O., Lindquist S.L., Ono B., Inge-Vechtomov S.G. & Liebman S.W. (1995) Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science, 268, 880–884. - PubMed
-
- Clark J.I. & Muchowski P.J. (2000) Small heatshock proteins and their potential role in human disease. Curr. Opin. Struct. Biol., 10, 52–59. - PubMed
-
- Daggett V. & Fersht A.R. (2003) Is there a unifying mechanism for protein folding? Trends Biochem. Sci., 28, 18–25. - PubMed
-
- Dinner A.R., Sali A., Smith L.J., Dobson C.M. & Karplus M. (2000) Understanding protein folding via free-energy surfaces from theory and experiment. Trends Biochem. Sci., 25, 331–339. - PubMed
-
- Farr G.W., Furtak K., Rowland M.B., Ranson N.A., Saibil H.R., Kirchhausen T. & Horwich A.L. (2000) Multivalent binding of nonnative substrate proteins by the chaperonin GroEL. Cell, 100, 561–573. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

