Purifying mRNAs with a high-affinity eIF4E mutant identifies the short 3' poly(A) end phenotype
- PMID: 12777618
- PMCID: PMC165825
- DOI: 10.1073/pnas.1232347100
Purifying mRNAs with a high-affinity eIF4E mutant identifies the short 3' poly(A) end phenotype
Abstract
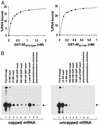
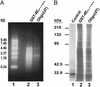
The use of DNA microarrays has revolutionized the manner in which mRNA populations are analyzed. One limitation of the current technology is that mRNAs are often purified on the basis of their 3' poly(A) ends, which can be extremely short or absent in some mRNAs. To circumvent this limitation, we have developed a procedure for the purification of eukaryotic mRNAs using a mutant version of the mRNA 5' cap-binding protein (eIF4E) with increased affinity for the m7GTP moiety of the cap. By using this procedure, we have compared the populations of mammalian mRNAs purified by oligo(dT) and 5' cap selection with oligonucleotide microarrays. This analysis has identified a subpopulation of mRNAs that are present with short 3' poly(A) ends at steady state and are missed or underrepresented after purification by oligo(dT). These mRNAs may respond to specific posttranscriptional control mechanisms such as cytoplasmic polyadenylation.
Figures

Similar articles
-
Efficient 5' cap-dependent RNA purification : use in identifying and studying subsets of RNA.Methods Mol Biol. 2008;419:147-60. doi: 10.1007/978-1-59745-033-1_10. Methods Mol Biol. 2008. PMID: 18369981
-
Characterization of human malaria parasite Plasmodium falciparum eIF4E homologue and mRNA 5' cap status.Mol Biochem Parasitol. 2007 Oct;155(2):146-55. doi: 10.1016/j.molbiopara.2007.07.003. Epub 2007 Jul 10. Mol Biochem Parasitol. 2007. PMID: 17692399
-
High affinity RNA for mammalian initiation factor 4E interferes with mRNA-cap binding and inhibits translation.RNA. 2005 Jan;11(1):77-89. doi: 10.1261/rna.7108205. RNA. 2005. PMID: 15611299 Free PMC article.
-
The translation of capped mRNAs has an absolute requirement for the central domain of eIF4G but not for the cap-binding initiation factor eIF4E.Cold Spring Harb Symp Quant Biol. 2001;66:377-87. doi: 10.1101/sqb.2001.66.377. Cold Spring Harb Symp Quant Biol. 2001. PMID: 12762040 Review. No abstract available.
-
Cap in hand: targeting eIF4E.Cell Cycle. 2009 Aug 15;8(16):2535-41. doi: 10.4161/cc.8.16.9301. Epub 2009 Aug 17. Cell Cycle. 2009. PMID: 19597330 Review.
Cited by
-
S-adenosylmethionine-dependent methyltransferase inhibitor DZNep blocks transcription and translation of SARS-CoV-2 genome with a low tendency to select for drug-resistant viral variants.Antiviral Res. 2022 Jan;197:105232. doi: 10.1016/j.antiviral.2021.105232. Epub 2021 Dec 29. Antiviral Res. 2022. PMID: 34968527 Free PMC article.
-
Determinants and implications of mRNA poly(A) tail size--does this protein make my tail look big?Semin Cell Dev Biol. 2014 Oct;34:24-32. doi: 10.1016/j.semcdb.2014.05.018. Epub 2014 Jun 5. Semin Cell Dev Biol. 2014. PMID: 24910447 Free PMC article. Review.
-
Global approaches for profiling transcription initiation.Cell Rep Methods. 2021 Sep 27;1(5):100081. doi: 10.1016/j.crmeth.2021.100081. Epub 2021 Sep 16. Cell Rep Methods. 2021. PMID: 34632443 Free PMC article. Review.
-
The arabidopsis RNA binding protein with K homology motifs, SHINY1, interacts with the C-terminal domain phosphatase-like 1 (CPL1) to repress stress-inducible gene expression.PLoS Genet. 2013;9(7):e1003625. doi: 10.1371/journal.pgen.1003625. Epub 2013 Jul 11. PLoS Genet. 2013. PMID: 23874224 Free PMC article.
-
Genome-wide characterization of methylguanosine-capped and polyadenylated small RNAs in the rice blast fungus Magnaporthe oryzae.Nucleic Acids Res. 2010 Nov;38(21):7558-69. doi: 10.1093/nar/gkq583. Epub 2010 Jul 21. Nucleic Acids Res. 2010. PMID: 20660015 Free PMC article.
References
-
- Tahara, S. M., Morgan, M. A. & Shatkin, A. J. (1981) J. Biol. Chem. 256, 7691–7694. - PubMed
-
- Grifo, J. A., Tahara, S. M., Morgan, M. A., Shatkin, A. J. & Merrick, W. C. (1983) J. Biol. Chem. 258, 5804–5810. - PubMed
-
- Pause, A., Belsham G. J., Gingras, A. C., Donze, O., Lin, T. A., Lawrence, J. C., Jr., & Sonenberg, N. (1994) Nature 371, 747–748. - PubMed
-
- Mothe-Satney, I., Brunn, G. J., McMahon, L. P., Capaldo, C. T., Abraham, R. T. & Lawrence, J. C., Jr., (2000) J. Biol. Chem. 275, 33836–33843. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

