Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin
- PMID: 12782659
- PMCID: PMC196073
- DOI: 10.1101/gad.1056603
Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin
Abstract
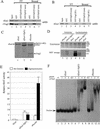
Drosophila Enhancer of Polycomb, E(Pc), is a suppressor of position-effect variegation and an enhancer of both Polycomb and trithorax mutations. A homologous yeast protein, Epl1, is a subunit of the NuA4 histone acetyltransferase complex. Epl1 depletion causes cells to accumulate in G2/M and global loss of acetylated histones H4 and H2A. In relation to the Drosophila protein, mutation of Epl1 suppresses gene silencing by telomere position effect. Epl1 protein is found in the NuA4 complex and a novel highly active smaller complex named Piccolo NuA4 (picNuA4). The picNuA4 complex contains Esa1, Epl1, and Yng2 as subunits and strongly prefers chromatin over free histones as substrate. Epl1 conserved N-terminal domain bridges Esa1 and Yng2 together, stimulating Esa1 catalytic activity and enabling acetylation of chromatin substrates. A recombinant picNuA4 complex shows characteristics similar to the native complex, including strong chromatin preference. Cells expressing only the N-terminal half of Epl1 lack NuA4 HAT activity, but possess picNuA4 complex and activity. These results indicate that the essential aspect of Esa1 and Epl1 resides in picNuA4 function. We propose that picNuA4 represents a nontargeted histone H4/H2A acetyltransferase activity responsible for global acetylation, whereas the NuA4 complex is recruited to specific genomic loci to perturb locally the dynamic acetylation/deacetylation equilibrium.
Figures
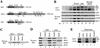




References
-
- Adams A., Gottschling, D.E., Kaiser, C.A., and Stearns, T. 1997. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Balasubramanian R., Pray-Grant, M.G., Selleck, W., Grant, P.A., and Tan, S. 2002. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 277: 7989–7995. - PubMed
-
- Bird A.W., Yu, D.Y., Pray-Grant, M.G., Qiu, Q., Harmon, K.E., Megee, P.C., Grant, P.A., Smith, M.M., and Christman, M.F. 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419: 411–415. - PubMed
-
- Brown C.E., Howe, L., Sousa, K., Alley, S.C., Carrozza, M.J., Tan, S., and Workman, J.L. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292: 2333–2337. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
