Neuroglian, Gliotactin, and the Na+/K+ ATPase are essential for septate junction function in Drosophila
- PMID: 12782686
- PMCID: PMC2172966
- DOI: 10.1083/jcb.200212054
Neuroglian, Gliotactin, and the Na+/K+ ATPase are essential for septate junction function in Drosophila
Abstract
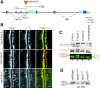
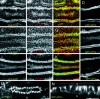
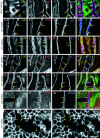
One essential function of epithelia is to form a barrier between the apical and basolateral surfaces of the epithelium. In vertebrate epithelia, the tight junction is the primary barrier to paracellular flow across epithelia, whereas in invertebrate epithelia, the septate junction (SJ) provides this function. In this study, we identify new proteins that are required for a functional paracellular barrier in Drosophila. In addition to the previously known components Coracle (COR) and Neurexin (NRX), we show that four other proteins, Gliotactin, Neuroglian (NRG), and both the alpha and beta subunits of the Na+/K+ ATPase, are required for formation of the paracellular barrier. In contrast to previous reports, we demonstrate that the Na pump is not localized basolaterally in epithelial cells, but instead is concentrated at the SJ. Data from immunoprecipitation and somatic mosaic studies suggest that COR, NRX, NRG, and the Na+/K+ ATPase form an interdependent complex. Furthermore, the observation that NRG, a Drosophila homologue of vertebrate neurofascin, is an SJ component is consistent with the notion that the invertebrate SJ is homologous to the vertebrate paranodal SJ. These findings have implications not only for invertebrate epithelia and barrier functions, but also for understanding of neuron-glial interactions in the mammalian nervous system.
Figures



References
-
- Ariyasu, R.G., and M.H. Ellisman. 1987. The distribution of (Na+/K+)ATPase is continuous along the axolemma of unensheathed axons from spinal roots of ‘dystrophic’ mice. J. Neurocytol. 16:239–248. - PubMed
-
- Arroyo, E.J., and S.S. Scherer. 2000. On the molecular architecture of myelinated fibers. Histochem. Cell Biol. 113:1–18. - PubMed
-
- Auld, V.J., R.D. Fetter, K. Broadie, and C.S. Goodman. 1995. Gliotactin, a novel transmembrane protein on peripheral glia, is required to form the blood-nerve barrier in Drosophila. Cell. 81:757–767. - PubMed
-
- Baumann, O. 2001. Posterior midgut epithelial cells differ in their organization of the membrane skeleton from other Drosophila epithelia. Exp. Cell Res. 270:176–187. - PubMed
-
- Baumgartner, S., J.T. Littleton, K. Broadie, M.A. Bhat, R. Harbecke, J.A. Lengyel, R. Chiquet-Ehrismann, A. Prokop, and H.J. Bellen. 1996. A Drosophila neurexin is required for septate junction and blood-nerve barrier formation and function. Cell. 87:1059–1068. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

