Human cytomegalovirus glycoprotein UL16 causes intracellular sequestration of NKG2D ligands, protecting against natural killer cell cytotoxicity
- PMID: 12782710
- PMCID: PMC2193902
- DOI: 10.1084/jem.20022059
Human cytomegalovirus glycoprotein UL16 causes intracellular sequestration of NKG2D ligands, protecting against natural killer cell cytotoxicity
Abstract
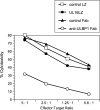
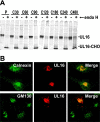
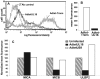
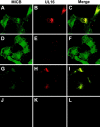
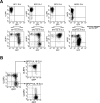
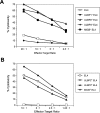
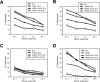
The activating receptor, NKG2D, is expressed on a variety of immune effector cells and recognizes divergent families of major histocompatibility complex (MHC) class I-related ligands, including the MIC and ULBP proteins. Infection, stress, or transformation can induce NKG2D ligand expression, resulting in effector cell activation and killing of the ligand-expressing target cell. The human cytomegalovirus (HCMV) membrane glycoprotein, UL16, binds to three of the five known ligands for human NKG2D. UL16 is retained in the endoplasmic reticulum and cis-Golgi apparatus of cells and causes MICB to be similarly retained and stabilized within cells. Coexpression of UL16 markedly reduces cell surface levels of MICB, ULBP1, and ULBP2, and decreases susceptibility to natural killer cell-mediated cytotoxicity. Domain swapping experiments demonstrate that the transmembrane and cytoplasmic domains of UL16 are important for intracellular retention of UL16, whereas the ectodomain of UL16 participates in down-regulation of NKG2D ligands. The intracellular sequestration of NKG2D ligands by UL16 represents a novel HCMV immune evasion mechanism to add to the well-documented viral strategies directed against antigen presentation by classical MHC molecules.
Figures



References
-
- Britt, W., and C. Alford. 1996. Cytomegalovirus. Field's Virology. B.N. Field, D.M. Knipe, and P.M. Howley, editors. Lippincott-Raven, Philadelphia, PA. 2493–2524.
-
- Tortorella, D., B.E. Gewurz, M.H. Furman, D.J. Schust, and H.L. Ploegh. 2000. Viral subversion of the immune system. Annu. Rev. Immunol. 18:861–926. - PubMed
-
- McFadden, G., and D.C. Johnson. 2002. Viral immune evasion. Immunology of Infectious Diseases. S.H.E. Kaufmann, A. Sher, and R. Ahmed, editors. American Society for Microbiology, Washington, D.C. 357–371.
-
- Tomazin, R., J. Boname, N.R. Hegde, D.M. Lewinsohn, Y. Altschuler, T.R. Jones, P. Cresswell, J.A. Nelson, S.R. Riddell, and D.C. Johnson. 1999. Cytomegalovirus US2 destroys two components of the MHC class II pathway, preventing recognition by CD4+ T cells. Nat. Med. 5:1039–1043. - PubMed
-
- Hegde, N.R., R.A. Tomazin, T.W. Wisner, C. Dunn, J.M. Boname, D.M. Lewinsohn, and D.C. Johnson. 2002. Inhibition of HLA-DR assembly, transport, and loading by human cytomegalovirus glycoprotein US3: a novel mechanism for evading major histocompatibility complex class II antigen presentation. J. Virol. 76:10929–10941. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

