PmSUC3: characterization of a SUT2/SUC3-type sucrose transporter from Plantago major
- PMID: 12782730
- PMCID: PMC156373
- DOI: 10.1105/tpc.010967
PmSUC3: characterization of a SUT2/SUC3-type sucrose transporter from Plantago major
Abstract
Higher plants possess medium-sized gene families that encode plasma membrane-localized sucrose transporters. For several plant species, it has been shown that at least one of these genes (e.g., AtSUC3 in Arabidopsis and LeSUT2 in tomato) differs from all other family members in several features, such as the length of the open reading frame, the number of introns, and the codon usage bias. For these reasons, and because two of these proteins did not rescue a yeast mutant defective in sucrose utilization, it had been speculated that this subgroup of transporters might have sensor functions. Here, we describe the detailed functional characterization and cellular localization of PmSUC3, the orthologous transporter from the Plantago major transporter family. The PmSUC3 protein is localized in the sieve elements of the Plantago phloem and mediates the energy-dependent transport of sucrose and maltose. In contrast to the situation in solanaceous plants, PmSUC3 is not colocalized with PmSUC2, the source-specific, phloem-loading sucrose transporter of Plantago. Moreover, PmSUC3 also was identified in sieve elements of sink leaves and in several nonphloem cells and tissues. Arguments for and against a potential sensor function for this type of sucrose transporter are presented, and the role of this type of transporter in the regulation of sucrose fluxes is discussed.
Figures


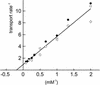
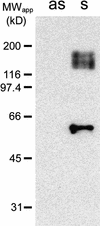
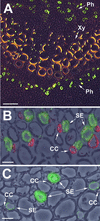

References
-
- Aoki, N., Hirose, T., Scofield, G.N., Whitfeld, P.R., and Furbank, R.T. (2003). The sucrose transporter gene family in rice. Plant Cell Physiol. 44, 223–232. - PubMed
-
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

