PCR analysis in archival postmortem tissues
- PMID: 12782767
- PMCID: PMC1187316
- DOI: 10.1136/mp.56.3.184
PCR analysis in archival postmortem tissues
Abstract
Background: Formalin fixed and paraffin wax embedded tissues of necropsy origin are an important source for molecular analysis especially in rare diseases, neuropathology, or molecular epidemiology studies. Because of DNA degradation, only short sequences can be amplified from this type of tissue, very often less than 100 bases. This poses problems because studies on polymorphism and mutations occurring in large genes often require the analysis of long sequences.
Methods: The development of a simple treatment to obtain longer fragments of DNA for the analysis of archival postmortem paraffin wax embedded tissues.
Results: It was possible to amplify longer sequences ranging up to 300 bases from postmortem tissues, with no modification to the usual DNA extraction procedures. To obtain longer stretches of DNA, a pre-PCR restoration treatment was required, by filling single strand breaks, followed by a vigorous denaturation step.
Conclusions: The development of this simple treatment allowed the analysis of longer fragments of DNA obtained from archival postmortem paraffin wax embedded tissues.
Figures
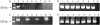

References
-
- Gressens P, Langston C, Mitchell WJ, et al. Detection of viral DNA in neonatal herpes encephalitis autopsy tissues by solution-phase PCR: comparison with pathology and immunohistochemistry. Brain Pathol 1993;3:237–50. - PubMed
-
- Stanta G, Croce LS, Bonin S, et al. Cohort effect of HCV infection in liver cirrhosis assessed by a 25 year study. J Clin Virol 2000;17:51–6. - PubMed
-
- Lehmann U, Kreipe H. Real-time PCR analysis of DNA and RNA extracted from formalin-fixed and paraffin-embedded biopsies. Methods 2001;25:409–18. - PubMed
-
- Voet D, Voet JG, Pratt CW. Nucleic acid structure. In: Fundamentals of biochemistry. New York: John Wiley and Sons, 1999:739–42.
-
- Ninet B, Rutschmann O, Burkhardt K, et al. Detection of mycobacterial nucleic acids by polymerase chain reaction in fixed tissue specimens of patients with human immunodeficiency virus infection. Swiss HIV cohort study. Diagn Mol Pathol 1999;8:145–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
