Allele-specific silencing of dominant disease genes
- PMID: 12782788
- PMCID: PMC165852
- DOI: 10.1073/pnas.1231012100
Allele-specific silencing of dominant disease genes
Abstract
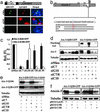
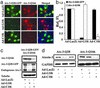
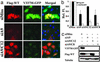
Small interfering RNA (siRNA) holds therapeutic promise for silencing dominantly acting disease genes, particularly if mutant alleles can be targeted selectively. In mammalian cell models we demonstrate that allele-specific silencing of disease genes with siRNA can be achieved by targeting either a linked single-nucleotide polymorphism (SNP) or the disease mutation directly. For a polyglutamine neurodegenerative disorder in which we first determined that selective targeting of the disease-causing CAG repeat is not possible, we took advantage of an associated SNP to generate siRNA that exclusively silenced the mutant Machado-Joseph disease/spinocerebellar ataxia type 3 allele while sparing expression of the WT allele. Allele-specific suppression was accomplished with all three approaches currently used to deliver siRNA: in vitro-synthesized duplexes as well as plasmid and viral expression of short hairpin RNA. We further optimized siRNA to specifically target a missense Tau mutation, V337M, that causes frontotemporal dementia. These studies establish that siRNA can be engineered to silence disease genes differing by a single nucleotide and highlight a key role for SNPs in extending the utility of siRNA in dominantly inherited disorders.
Figures

Similar articles
-
Targeting Alzheimer's disease genes with RNA interference: an efficient strategy for silencing mutant alleles.Nucleic Acids Res. 2004 Jan 30;32(2):661-8. doi: 10.1093/nar/gkh208. Print 2004. Nucleic Acids Res. 2004. PMID: 14754988 Free PMC article.
-
Allele-specific RNA silencing of mutant ataxin-3 mediates neuroprotection in a rat model of Machado-Joseph disease.PLoS One. 2008 Oct 8;3(10):e3341. doi: 10.1371/journal.pone.0003341. PLoS One. 2008. PMID: 18841197 Free PMC article.
-
Molecular and clinical correlations in spinocerebellar ataxia type 3 and Machado-Joseph disease.Ann Neurol. 1995 Jul;38(1):68-72. doi: 10.1002/ana.410380113. Ann Neurol. 1995. PMID: 7611728
-
Dominantly inherited ataxias: lessons learned from Machado-Joseph disease/spinocerebellar ataxia type 3.Semin Neurol. 2007 Apr;27(2):133-42. doi: 10.1055/s-2007-971172. Semin Neurol. 2007. PMID: 17390258 Review.
-
Therapy for dominant inherited diseases by allele-specific RNA interference: successes and pitfalls.Curr Gene Ther. 2015;15(5):503-10. doi: 10.2174/1566523215666150812115730. Curr Gene Ther. 2015. PMID: 26264709 Review.
Cited by
-
RNA interference technologies and therapeutics: from basic research to products.BioDrugs. 2009;23(5):305-32. doi: 10.2165/11318190-000000000-00000. BioDrugs. 2009. PMID: 19754220 Free PMC article. Review.
-
Ataxin-3 protein and RNA toxicity in spinocerebellar ataxia type 3: current insights and emerging therapeutic strategies.Mol Neurobiol. 2014 Jun;49(3):1513-31. doi: 10.1007/s12035-013-8596-2. Epub 2013 Nov 29. Mol Neurobiol. 2014. PMID: 24293103 Free PMC article. Review.
-
RNAi-based drug design: considerations and future directions.Nat Rev Drug Discov. 2024 May;23(5):341-364. doi: 10.1038/s41573-024-00912-9. Epub 2024 Apr 3. Nat Rev Drug Discov. 2024. PMID: 38570694 Free PMC article. Review.
-
Identification of Novel Fibrosis Modifiers by In Vivo siRNA Silencing.Mol Ther Nucleic Acids. 2017 Jun 16;7:314-323. doi: 10.1016/j.omtn.2017.04.014. Epub 2017 Apr 20. Mol Ther Nucleic Acids. 2017. PMID: 28624207 Free PMC article.
-
A Tandem Oligonucleotide Approach for SNP-Selective RNA Degradation Using Modified Antisense Oligonucleotides.PLoS One. 2015 Nov 6;10(11):e0142139. doi: 10.1371/journal.pone.0142139. eCollection 2015. PLoS One. 2015. PMID: 26544037 Free PMC article.
References
-
- McManus, M. T. & Sharp, P. A. (2002) Nat. Rev. Genet. 3, 737-747. - PubMed
-
- Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494-498. - PubMed
-
- Brummelkamp, T. R., Bernards, R. & Agami, R. (2002) Science 296, 550-553. - PubMed
-
- McCaffrey, A. P., Meuse, L., Pham, T. T., Conklin, D. S., Hannon, G. J. & Kay, M. A. (2002) Nature 418, 38-39. - PubMed
-
- Xia, H., Mao, Q., Paulson, H. L. & Davidson, B. L. (2002) Nat. Biotechnol. 20, 1006-1010. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

