Periodic repression by the bHLH factor Hes7 is an essential mechanism for the somite segmentation clock
- PMID: 12783854
- PMCID: PMC196074
- DOI: 10.1101/gad.1092303
Periodic repression by the bHLH factor Hes7 is an essential mechanism for the somite segmentation clock
Abstract
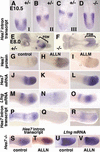
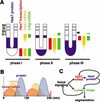
Hes7, a bHLH gene essential for somitogenesis, displays cyclic expression of mRNA in the presomitic mesoderm (PSM). Here, we show that Hes7 protein is also expressed in a dynamic manner, which depends on proteasome-mediated degradation. Spatial comparison revealed that Hes7 and Lunatic fringe (Lfng) transcription occurs in the Hes7 protein-negative domains. Furthermore, Hes7 and Lfng transcription is constitutively up-regulated in the absence of Hes7 protein and down-regulated by stabilization of Hes7 protein. Thus, periodic repression by Hes7 protein is critical for the cyclic transcription of Hes7 and Lfng, and this negative feedback represents a molecular basis for the segmentation clock.
Figures


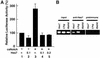
References
-
- Aulehla A. and Johnson, R.L. 1999. Dynamic expression of Lunatic fringe suggests a link between notch signaling and an autonomous cellular oscillator driving somite segmentation. Dev. Biol. 207: 49–61. - PubMed
-
- Aulehla A., Wehrle, C., Brand-Saberi, B., Kemler, R., Gossler, A., Kanzler, B., and Herrmann, B.G. 2003. Wnt3a plays a major role in the segmentation clock controlling somitogenesis. Dev. Cell 4: 395–406. - PubMed
-
- Bessho Y., Miyoshi, G., Sakata, R., and Kageyama, R. 2001a. Hes7: A bHLH-type repressor gene regulated by Notch and expressed in the presomitic mesoderm. Genes Cells 6: 175–185. - PubMed
-
- Cole S.E., Levorse, J.M., Tilghman, S.M., and Vogt, T.F. 2002. Clock regulatory elements control cyclic expression of Lunatic fringe during somitogenesis. Dev. Cell 3: 75–84. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
