Enhanced mercury biosorption by bacterial cells with surface-displayed MerR
- PMID: 12788714
- PMCID: PMC161548
- DOI: 10.1128/AEM.69.6.3176-3180.2003
Enhanced mercury biosorption by bacterial cells with surface-displayed MerR
Abstract
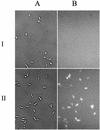
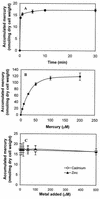
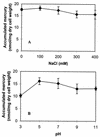
The metalloregulatory protein MerR, which exhibits high affinity and selectivity toward mercury, was exploited for the construction of microbial biosorbents specific for mercury removal. Whole-cell sorbents were constructed with MerR genetically engineered onto the surface of Escherichia coli cells by using an ice nucleation protein anchor. The presence of surface-exposed MerR on the engineered strains enabled sixfold-higher Hg(2+) biosorption than that found in the wild-type JM109 cells. Hg(2+) binding via MerR was very specific, with no observable decline even in the presence of 100-fold excess Cd(2+) and Zn(2+). The Hg(2+) binding property of the whole-cell sorbents was also insensitive to different ionic strengths, pHs, and the presence of metal chelators. Since metalloregulatory proteins are currently available for a wide variety of toxic heavy metals, our results suggest that microbial biosorbents overexpressing metalloregulatory proteins may be used similarly for the cleanup of other important heavy metals.
Figures

References
-
- Bae, W., W. Chen, A. Mulchandani, and R. Mehra. 2000. Enhanced bioaccumulation of heavy metals by bacterial cells displaying synthetic phytochelatins. Biotechnol. Bioeng. 70:518-523. - PubMed
-
- Bae, W., A. Mulchandani, and W. Chen. 2002. Cell surface display of synthetic phytochelatins using ice nucleation protein for enhanced heavy metal bioaccumulation. J. Inorg. Biochem. 88:223-227. - PubMed
-
- Bontidean, I., C. Berggren, G. Johansson, E. Csorgi, B. Mattiasson, J. R. Lloyd, K. J. Jakeman, and N. L. Brown. 1998. Detection of heavy metal ions at femtomolar levels using protein-based biosensors. Anal. Chem. 70:4162-4169. - PubMed
-
- Brown, W. C., and J. L. Campbell. 1993. A new cloning vector and expression strategy for genes encoding proteins toxic to Escherichia coli. Gene 127:99-103. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

