Detection and enumeration of aromatic oxygenase genes by multiplex and real-time PCR
- PMID: 12788736
- PMCID: PMC161477
- DOI: 10.1128/AEM.69.6.3350-3358.2003
Detection and enumeration of aromatic oxygenase genes by multiplex and real-time PCR
Abstract
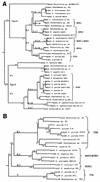
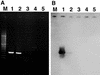
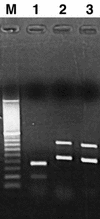
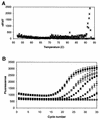
Our abilities to detect and enumerate pollutant-biodegrading microorganisms in the environment are rapidly advancing with the development of molecular genetic techniques. Techniques based on multiplex and real-time PCR amplification of aromatic oxygenase genes were developed to detect and quantify aromatic catabolic pathways, respectively. PCR primer sets were identified for the large subunits of aromatic oxygenases from alignments of known gene sequences and tested with genetically well-characterized strains. In all, primer sets which allowed amplification of naphthalene dioxygenase, biphenyl dioxygenase, toluene dioxygenase, xylene monooxygenase, phenol monooxygenase, and ring-hydroxylating toluene monooxygenase genes were identified. For each primer set, the length of the observed amplification product matched the length predicted from published sequences, and specificity was confirmed by hybridization. Primer sets were grouped according to the annealing temperature for multiplex PCR permitting simultaneous detection of various genotypes responsible for aromatic hydrocarbon biodegradation. Real-time PCR using SYBR green I was employed with the individual primer sets to determine the gene copy number. Optimum polymerization temperatures for real-time PCR were determined on the basis of the observed melting temperatures of the desired products. When a polymerization temperature of 4 to 5 degrees C below the melting temperature was used, background fluorescence signals were greatly reduced, allowing detection limits of 2 x 10(2) copies per reaction mixture. Improved in situ microbial characterization will provide more accurate assessment of pollutant biodegradation, enhance studies of the ecology of contaminated sites, and facilitate assessment of the impact of remediation technologies on indigenous microbial populations.
Figures
References
-
- Aoki, H., T. Kimura, H. Habe, H. Yumane, T. Kodama, and T. Omori. 1996. Cloning, nucleotide sequence, and characterization of the genes encoding enzymes involved in the degradation of cumene to 2-hydroxy-6-oxo-7-methylocta-2,4-dienoic acid in Pseudomonas fluorescens IP01. J. Ferment. Bioeng. 81:187-196.
-
- Asturias, J. A., E. Dìaz, and K. N. Timmis. 1995. The evolutionary relationship of biphenyl dioxygenase from gram-positive Rhodococcus globerulus P6 to multicomponent dioxygenase from gram-negative bacteria. Gene 156:11-18. - PubMed
-
- Boronin, A. M., T. V. Tsoi, I. A. Kosheleva, M. U. Arinbasarov, and V. M. Adanin. 1989. Cloning of Pseudomonas putida genes responsible for the primary stages of oxidation of naphthalene in Escherichia coli cells. Genetika 25:226-237. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

