Use of PCR for direct detection of Campylobacter species in bovine feces
- PMID: 12788747
- PMCID: PMC161499
- DOI: 10.1128/AEM.69.6.3435-3447.2003
Use of PCR for direct detection of Campylobacter species in bovine feces
Abstract
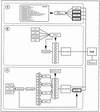
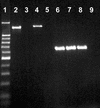
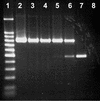
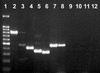
This study reports on the use of PCR to directly detect and distinguish Campylobacter species in bovine feces without enrichment. Inhibitors present in feces are a major obstacle to using PCR to detect microorganisms. The QIAamp DNA stool minikit was found to be an efficacious extraction method, as determined by the positive amplification of internal control DNA added to bovine feces before extraction. With nested or seminested multiplex PCR, Campylobacter coli, C. fetus, C. hyointestinalis, and C. jejuni were detected in all fecal samples inoculated at approximately 10(4) CFU g(-1), and 50 to 83% of the samples inoculated at approximately 10(3) CFU g(-1) were positive. At approximately 10(2) CFU g(-1), C. fetus, C. hyointestinalis, and C. jejuni (17 to 50% of the samples) but not C. coli were detected by PCR. From uninoculated bovine feces, a total of 198 arbitrarily selected isolates of Campylobacter were recovered on four commonly used isolation media incubated at three temperatures. The most frequently isolated taxa were C. jejuni (152 isolates) and C. lanienae (42 isolates), but isolates of C. fetus subsp. fetus, Arcobacter butzleri, and A. skirrowii also were recovered (</=2 isolates per taxon). Considerable variability was observed in the frequency of isolation of campylobacters among the four media and three incubation temperatures tested. With genus-specific primers, Campylobacter DNA was detected in 75% of the fecal samples, representing an 8% increase in sensitivity relative to that obtained with microbiological isolation across the four media and three incubation temperatures tested. With nested primers, C. jejuni and C. lanienae were detected in 25 and 67% of the samples, respectively. In no instance was DNA from either C. coli, C. fetus, or C. hyointestinalis detected in uninoculated bovine feces. PCR was more sensitive than isolation on microbiological media for detecting C. lanienae (17%) but not C. jejuni. Campylobacters are a diverse and fastidious group of bacteria, and the development of direct PCR not only will increase the understanding of Campylobacter species diversity and their frequency of occurrence in feces but also will enhance the knowledge of their role in the gastrointestinal tract of livestock and of the factors that influence shedding.
Figures


References
-
- Atabay, H. I., and J. E. L. Corry. 1998. The isolation and prevalence of campylobacters from dairy cattle using a variety of methods. J. Appl. Microbiol. 84:733-740. - PubMed
-
- Ballagi-Pordany, A., and S. Belak. 1996. The use of mimics as internal standards to avoid false negatives in diagnostic PCR. Mol. Cell. Probes 10:159-164. - PubMed
-
- Bastyns, K., S. Chapelle, P. Vandamme, H. Goossens, and R. de Wachter. 1994. Species-specific detection of campylobacters important in veterinary medicine by PCR amplification of 23S rDNA areas. Syst. Appl. Microbiol. 17:563-568.
-
- Baylis, C. L., S. MacPhee, K. W. Martin, T. J. Humphrey, and R. P. Betts. 2000. Comparison of three enrichment media for the isolation of Campylobacter spp. from foods. J. Appl. Microbiol. 89:884-891. - PubMed
-
- Busato, A., D. Hofer, T. Lentze, C. Gaillard, and A. Burnens. 1999. Prevalence and infection risks of zoonotic enteropathogenic bacteria in Swiss cow-calf farms. Vet. Microbiol. 69:251-263. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

