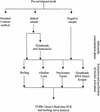
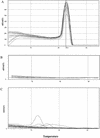
Evaluation of DNA extraction methods for use in combination with SYBR green I real-time PCR to detect Salmonella enterica serotype enteritidis in poultry
- PMID: 12788750
- PMCID: PMC161507
- DOI: 10.1128/AEM.69.6.3456-3461.2003
Evaluation of DNA extraction methods for use in combination with SYBR green I real-time PCR to detect Salmonella enterica serotype enteritidis in poultry
Abstract
The objective of this study was to develop a rapid, reproducible, and robust method for detecting Salmonella enterica serotype Enteritidis in poultry samples. First, for the extraction and purification of DNA from the preenrichment culture, four methods (boiling, alkaline lysis, Nucleospin, and Dynabeads DNA Direct System I) were compared. The most effective method was then combined with a real-time PCR method based on the double-stranded DNA binding dye SYBR Green I used with the ABI Prism 7700 system. The specificity of the reaction was determined by the melting temperature (T(m)) of the amplicon obtained. The experiments were conducted both on samples of chicken experimentally contaminated with serotype Enteritidis and on commercially available poultry samples, which were also used for comparisons with the standard cultural method (i.e., ISO 6579/2001). The results of comparisons among the four DNA extraction methods showed significant differences except for the results from the boiling and Nucleospin methods (the two methods that produced the lowest threshold cycles). Boiling was selected as the preferred extraction method because it is the simplest and most rapid. This method was then combined with SYBR Green I real-time PCR, using primers SEFA-1 and SEFA-2. The specificity of the reaction was confirmed by the T(m), which was consistently specific for the amplicon obtained; the mean peak T(m) obtained with curves specific for serotype Enteritidis was 82.56 +/- 0.22 degrees C. The standard curve constructed using the mean threshold cycle and various concentrations of serotype Enteritidis (ranging from 10(3) to 10(8) CFU/ml) showed good linearity (R(2) = 0.9767) and a sensitivity limit of less than 10(3) CFU/ml. The results of this study demonstrate that the SYBR Green I real-time PCR constitutes an effective and easy-to-perform method for detecting serotype Enteritidis in poultry samples.
Figures
Similar articles
-
Development of a cell culture method to isolate and enrich Salmonella enterica serotype enteritidis from shell eggs for subsequent detection by real-time PCR.Appl Environ Microbiol. 2009 Aug;75(16):5321-7. doi: 10.1128/AEM.02422-08. Epub 2009 Jun 26. Appl Environ Microbiol. 2009. PMID: 19561188 Free PMC article.
-
Loop-Mediated Isothermal Amplification of the sefA Gene for Rapid Detection of Salmonella Enteritidis and Salmonella Gallinarum in Chickens.Foodborne Pathog Dis. 2016 Apr;13(4):177-81. doi: 10.1089/fpd.2015.2082. Epub 2016 Feb 3. Foodborne Pathog Dis. 2016. PMID: 26840841
-
Optimization of rapid Salmonella enterica detection in liquid whole eggs by SYBR green I-based real-time reverse transcriptase-polymerase chain reaction.Foodborne Pathog Dis. 2011 Apr;8(4):527-34. doi: 10.1089/fpd.2010.0721. Epub 2011 Mar 7. Foodborne Pathog Dis. 2011. PMID: 21381900
-
Detection of Salmonella enteritidis in pooled poultry environmental samples using a serotype-specific real-time-polymerase chain reaction assay.Avian Dis. 2013 Mar;57(1):22-8. doi: 10.1637/10279-061312-Reg.1. Avian Dis. 2013. PMID: 23678725
-
The Significance and Importance of dPCR, qPCR, and SYBR Green PCR Kit in the Detection of Numerous Diseases.Curr Pharm Des. 2024;30(3):169-179. doi: 10.2174/0113816128276560231218090436. Curr Pharm Des. 2024. PMID: 38243947 Review.
Cited by
-
Persistence of bacterial indicators and zoonotic pathogens in contaminated cattle wastes.BMC Microbiol. 2016 May 20;16:87. doi: 10.1186/s12866-016-0705-8. BMC Microbiol. 2016. PMID: 27206734 Free PMC article.
-
Gastrointestinal tract distribution of Salmonella enteritidis in orally infected mice with a species-specific fluorescent quantitative polymerase chain reaction.World J Gastroenterol. 2007 Dec 28;13(48):6568-74. doi: 10.3748/wjg.v13.i48.6568. World J Gastroenterol. 2007. PMID: 18161929 Free PMC article.
-
Multiplex real-time PCR for detection of Staphylococcus aureus, mecA and Panton-Valentine Leukocidin (PVL) genes from selective enrichments from animals and retail meat.PLoS One. 2014 May 21;9(5):e97617. doi: 10.1371/journal.pone.0097617. eCollection 2014. PLoS One. 2014. PMID: 24849624 Free PMC article.
-
Transmission and Toxigenic Potential of Vibrio cholerae in Hilsha Fish (Tenualosa ilisha) for Human Consumption in Bangladesh.Front Microbiol. 2018 Feb 20;9:222. doi: 10.3389/fmicb.2018.00222. eCollection 2018. Front Microbiol. 2018. PMID: 29515532 Free PMC article.
-
Pathogenic potential and antibiotic resistance of Yersinia enterocolitica, a foodborne pathogen limited to swine tonsils in a pork production chain from Southern Brazil.Braz J Microbiol. 2021 Dec;52(4):2335-2342. doi: 10.1007/s42770-021-00591-3. Epub 2021 Aug 18. Braz J Microbiol. 2021. PMID: 34406639 Free PMC article.
References
-
- Aarts, H. J., R. G. Joosten, M. H. Henkens, H. Stegeman, and A. H. van Hoek. 2001. Rapid duplex PCR assay for the detection of pathogenic Yersinia enterocolitica strains. J. Microbiol. Methods 47:209-217. - PubMed
-
- Alterkruse, S. F., L. K. Tollefson, and K. Bögel. 1993. Control strategies for Salmonella enteritidis in five countries. Food Control 4:10-16.
-
- Baumler, A. J., B. M. Hargis, and R. M. Tsolis. 2000. Tracing the origins of Salmonella outbreaks. Science 287:50-52. [Online.] http://www.sciencemag.org/cgi/content/full/287/5450/50. - PubMed
-
- Chen, S., A. Yee, M. Griffiths, C. Larkin, C. T. Yamashiro, R. Behari, C. Paszko-Kolva, K. Rahn, and S. A. De Grandis. 1997. The evaluation of a fluorogenic polymerase chain reaction assay for the detection of Salmonella species in food commodities. Int. J. Food Microbiol. 35:239-250. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases