A docking site in MKK4 mediates high affinity binding to JNK MAPKs and competes with similar docking sites in JNK substrates
- PMID: 12788955
- PMCID: PMC3017503
- DOI: 10.1074/jbc.M304229200
A docking site in MKK4 mediates high affinity binding to JNK MAPKs and competes with similar docking sites in JNK substrates
Abstract
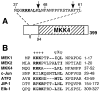
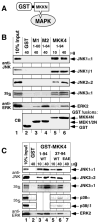
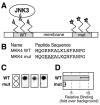
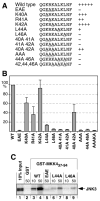
Specific docking interactions between MAPKs and their activating MAPK kinases (MKKs or MEKs) are crucial for efficient and accurate signal transmission. Here, we report the identification of a MAPK-docking site, or "D-site," in the N terminus of human MKK4/JNKK1. This docking site conforms to the consensus sequence for known D-sites in other MKKs and contains the first of the two cleavage sites for anthrax lethal factor protease that have been found in the N terminus of MKK4. This docking site was both necessary and sufficient for the high affinity binding of the MAPKs JNK1, JNK2, JNK3, p38 alpha, and p38 beta to MKK4. Mutations that altered conserved residues in this docking site reduced JNK/p38 binding. In addition, a peptide version of this docking site, as well as a peptide version of the JNK-binding site of the JIP-1 scaffold protein, inhibited both MKK4/JNK binding and MKK4-mediated phosphorylation of JNK1. These same peptides also inhibited JNK2-mediated phosphorylation of c-Jun and ATF2, suggesting that transcription factors, MKK4, and the JIP scaffold compete for docking to JNK. Finally, the selectivity of the MKK4, MEK1, and MEK2 D-sites for JNK versus ERK was quantified. The MEK1 and MEK2 D-sites displayed a strong selectivity for their cognate MAPK (ERK2) versus a non-cognate MAPK (JNK). In contrast, the MKK4 D-site exhibited only limited selectivity for JNK versus ERK.
Figures




Similar articles
-
A conserved docking site in MEKs mediates high-affinity binding to MAP kinases and cooperates with a scaffold protein to enhance signal transmission.J Biol Chem. 2001 Mar 30;276(13):10374-86. doi: 10.1074/jbc.M010271200. Epub 2000 Dec 28. J Biol Chem. 2001. PMID: 11134045 Free PMC article.
-
Selectivity of docking sites in MAPK kinases.J Biol Chem. 2009 May 8;284(19):13165-73. doi: 10.1074/jbc.M900080200. Epub 2009 Feb 5. J Biol Chem. 2009. PMID: 19196711 Free PMC article.
-
Docking sites on mitogen-activated protein kinase (MAPK) kinases, MAPK phosphatases and the Elk-1 transcription factor compete for MAPK binding and are crucial for enzymic activity.Biochem J. 2003 Mar 15;370(Pt 3):1077-85. doi: 10.1042/BJ20021806. Biochem J. 2003. PMID: 12529172 Free PMC article.
-
Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases.Science. 2002 Dec 6;298(5600):1911-2. doi: 10.1126/science.1072682. Science. 2002. PMID: 12471242 Review.
-
Physiological roles of MKK4 and MKK7: insights from animal models.Biochim Biophys Acta. 2007 Aug;1773(8):1349-57. doi: 10.1016/j.bbamcr.2006.10.016. Epub 2006 Nov 10. Biochim Biophys Acta. 2007. PMID: 17157936 Review.
Cited by
-
Bacterial ubiquitin ligase engineered for small molecule and protein target identification.bioRxiv [Preprint]. 2025 Mar 22:2025.03.20.644192. doi: 10.1101/2025.03.20.644192. bioRxiv. 2025. PMID: 40166235 Free PMC article. Preprint.
-
Unfolded protein response in colorectal cancer.Cell Biosci. 2021 Jan 29;11(1):26. doi: 10.1186/s13578-021-00538-z. Cell Biosci. 2021. PMID: 33514437 Free PMC article. Review.
-
A conserved protein interaction network involving the yeast MAP kinases Fus3 and Kss1.J Cell Biol. 2004 Jan 19;164(2):267-77. doi: 10.1083/jcb.200310021. J Cell Biol. 2004. PMID: 14734536 Free PMC article.
-
Glucocorticoid receptor-JNK interaction mediates inhibition of the JNK pathway by glucocorticoids.EMBO J. 2003 Nov 17;22(22):6035-44. doi: 10.1093/emboj/cdg590. EMBO J. 2003. PMID: 14609950 Free PMC article.
-
Proteome-wide screening for mitogen-activated protein kinase docking motifs and interactors.Sci Signal. 2023 Jan 10;16(767):eabm5518. doi: 10.1126/scisignal.abm5518. Epub 2023 Jan 10. Sci Signal. 2023. PMID: 36626580 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

