A hospital-associated outbreak of Legionnaires' disease caused by Legionella pneumophila serogroup 1 is characterized by stable genetic fingerprinting but variable monoclonal antibody patterns
- PMID: 12791873
- PMCID: PMC156525
- DOI: 10.1128/JCM.41.6.2503-2508.2003
A hospital-associated outbreak of Legionnaires' disease caused by Legionella pneumophila serogroup 1 is characterized by stable genetic fingerprinting but variable monoclonal antibody patterns
Abstract
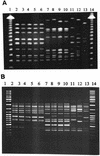
An outbreak of 18 pneumonia cases caused by Legionella pneumophila serogroup 1 occurred at a Swedish university hospital 1996 to 1999. Eight clinical isolates obtained by culture from the respiratory tract were compared to 20 environmental isolates from the hospital and to 21 epidemiologically unrelated isolates in Sweden, mostly from patients, by using pulsed-field gel electrophoresis (PFGE), amplified fragment length polymorphism analysis (AFLP), and monoclonal antibody (MAb) typing. All patients and most environmental isolates from the outbreak hospital belonged to the same genotypic cluster in both PFGE and AFLP. This genotype was distinctly different from other strains, including a cluster from a second hospital in a different part of the country. The MAb subtype of the outbreak clone was Knoxville except for three isolates that were Oxford. A variation in the MAb reactivity pattern was also found in a second genotypic cluster. These changes in the MAb reactivity pattern were due to the absence or presence of the lag-1 gene coding for an O-acetyltransferase that is responsible for expression of the lipopolysaccharide epitope recognized by MAb 3/1 of the Dresden Panel. In all MAb 3/1-positive strains, the lag-1 gene was present on a genetic element that was bordered by a direct repeat that showed a high degree of sequence homology. Due to this homology, the lag-1 gene region seemed to be an unstable element in the chromosome. MAb patterns are thus a valuable adjunct to genotyping methods in defining subgroups inside a genotypic cluster of L. pneumophila sg 1.
Figures



References
-
- Bernander, S., K. Jacobson, U. Zettersten, J. H. Helbig, and M. Lundholm. 2000. PFGE, AFLP, and MAb subtyping of Legionella pneumophila serogroup 1. Clin. Microbiol. Infect. 6(Suppl. 1):223.
-
- Colbourne, J. S., and P. J. Dennis. 1989. The ecology and survival of Legionella pneumophila. J. Institution Water Environ. Management 3:345-350.
-
- Dournon, E., W. F. Bibb, P. Rajagopalan, N. Desplaces, and R. M. McKinney. 1988. Monoclonal antibody reactivity as a virulence marker for Legionella pneumophila serogroup 1 strains. J. Infect. Dis. 157:496-501. - PubMed
-
- Drenning, S. D., J. E. Stout, J. R. Joly, and V. L. Yu. 2001. Unexpected similarity of pulsed-field gel electrophoresis patterns of unrelated clinical isolates of Legionella pneumophila, serogroup 1. J. Infect. Dis. 183:628-632. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

