X-ray structure of human acid-beta-glucosidase, the defective enzyme in Gaucher disease
- PMID: 12792654
- PMCID: PMC1326319
- DOI: 10.1038/sj.embor.embor873
X-ray structure of human acid-beta-glucosidase, the defective enzyme in Gaucher disease
Abstract
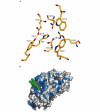
Gaucher disease, the most common lysosomal storage disease, is caused by mutations in the gene that encodes acid-beta-glucosidase (GlcCerase). Type 1 is characterized by hepatosplenomegaly, and types 2 and 3 by early or chronic onset of severe neurological symptoms. No clear correlation exists between the approximately 200 GlcCerase mutations and disease severity, although homozygosity for the common mutations N370S and L444P is associated with non- neuronopathic and neuronopathic disease, respectively. We report the X-ray structure of GlcCerase at 2.0 A resolution. The catalytic domain consists of a (beta/alpha)(8) TIM barrel, as expected for a member of the glucosidase hydrolase A clan. The distance between the catalytic residues E235 and E340 is consistent with a catalytic mechanism of retention. N370 is located on the longest alpha-helix (helix 7), which has several other mutations of residues that point into the TIM barrel. Helix 7 is at the interface between the TIM barrel and a separate immunoglobulin-like domain on which L444 is located, suggesting an important regulatory or structural role for this non-catalytic domain. The structure provides the possibility of engineering improved GlcCerase for enzyme-replacement therapy, and for designing structure-based drugs aimed at restoring the activity of defective GlcCerase.
Figures


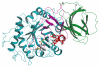
References
-
- Abrahams J.P. & Leslie A.G. (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D, 52, 30–42. - PubMed
-
- Berg-Fussman A., Grace M.E., Ioannou Y. & Grabowski G.A. (1993) Human acid β-glucosidase. N-glycosylation site occupancy and the effect of glycosylation on enzymatic activity. J. Biol. Chem., 268 14861–14866. - PubMed
-
- Beutler E. & Grabowski G.A. (2001) in The Metabolic and Molecular Bases of Inherited Disease (eds Scriver, C.R., Sly, W.S., Childs, B., Beaudet, A.L., Valle, D., Kinzler, K.W. & Vogelstein, B), 3635–3668. McGraw-Hill, Inc., New York, USA.
-
- Brunger A.T. et al. . (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr., 54, 905–921. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

