RNA processing and degradation in Bacillus subtilis
- PMID: 12794188
- PMCID: PMC156466
- DOI: 10.1128/MMBR.67.2.157-174.2003
RNA processing and degradation in Bacillus subtilis
Abstract
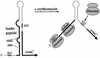
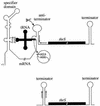
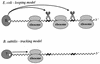
This review focuses on the enzymes and pathways of RNA processing and degradation in Bacillus subtilis, and compares them to those of its gram-negative counterpart, Escherichia coli. A comparison of the genomes from the two organisms reveals that B. subtilis has a very different selection of RNases available for RNA maturation. Of 17 characterized ribonuclease activities thus far identified in E. coli and B. subtilis, only 6 are shared, 3 exoribonucleases and 3 endoribonucleases. Some enzymes essential for cell viability in E. coli, such as RNase E and oligoribonuclease, do not have homologs in B. subtilis, and of those enzymes in common, some combinations are essential in one organism but not in the other. The degradation pathways and transcript half-lives have been examined to various degrees for a dozen or so B. subtilis mRNAs. The determinants of mRNA stability have been characterized for a number of these and point to a fundamentally different process in the initiation of mRNA decay. While RNase E binds to the 5' end and catalyzes the rate-limiting cleavage of the majority of E. coli RNAs by looping to internal sites, the equivalent nuclease in B. subtilis, although not yet identified, is predicted to scan or track from the 5' end. RNase E can also access cleavage sites directly, albeit less efficiently, while the enzyme responsible for initiating the decay of B. subtilis mRNAs appears incapable of direct entry. Thus, unlike E. coli, RNAs possessing stable secondary structures or sites for protein or ribosome binding near the 5' end can have very long half-lives even if the RNA is not protected by translation.
Figures

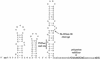


Similar articles
-
RNA degradation in Bacillus subtilis: an interplay of essential endo- and exoribonucleases.Mol Microbiol. 2012 Jun;84(6):1005-17. doi: 10.1111/j.1365-2958.2012.08072.x. Epub 2012 May 8. Mol Microbiol. 2012. PMID: 22568516 Review.
-
Decay of a model mRNA in Bacillus subtilis by a combination of RNase J1 5' exonuclease and RNase Y endonuclease activities.J Bacteriol. 2011 Nov;193(22):6384-6. doi: 10.1128/JB.05939-11. Epub 2011 Sep 9. J Bacteriol. 2011. PMID: 21908660 Free PMC article.
-
Small stable RNA maturation and turnover in Bacillus subtilis.Mol Microbiol. 2015 Jan;95(2):270-82. doi: 10.1111/mmi.12863. Epub 2014 Dec 19. Mol Microbiol. 2015. PMID: 25402410 Free PMC article.
-
Dynamic Membrane Localization of RNase Y in Bacillus subtilis.mBio. 2020 Feb 18;11(1):e03337-19. doi: 10.1128/mBio.03337-19. mBio. 2020. PMID: 32071272 Free PMC article.
-
Bacillus subtilis mRNA decay: new parts in the toolkit.Wiley Interdiscip Rev RNA. 2011 May-Jun;2(3):387-94. doi: 10.1002/wrna.66. Epub 2010 Dec 16. Wiley Interdiscip Rev RNA. 2011. PMID: 21957024 Review.
Cited by
-
Antisense RNA regulation by stable complex formation in the Enterococcus faecalis plasmid pAD1 par addiction system.J Bacteriol. 2004 Oct;186(19):6400-8. doi: 10.1128/JB.186.19.6400-6408.2004. J Bacteriol. 2004. PMID: 15375120 Free PMC article.
-
Role of Bacillus subtilis RNase J1 endonuclease and 5'-exonuclease activities in trp leader RNA turnover.J Biol Chem. 2008 Jun 20;283(25):17158-67. doi: 10.1074/jbc.M801461200. Epub 2008 Apr 29. J Biol Chem. 2008. PMID: 18445592 Free PMC article.
-
Transcriptional analysis of the cip-cel gene cluster from Clostridium cellulolyticum.J Bacteriol. 2006 Apr;188(7):2614-24. doi: 10.1128/JB.188.7.2614-2624.2006. J Bacteriol. 2006. PMID: 16547049 Free PMC article.
-
The bag or the spindle: the cell factory at the time of systems' biology.Microb Cell Fact. 2004 Nov 10;3(1):13. doi: 10.1186/1475-2859-3-13. Microb Cell Fact. 2004. PMID: 15537427 Free PMC article.
-
Stable PNPase RNAi silencing: its effect on the processing and adenylation of human mitochondrial RNA.RNA. 2008 Feb;14(2):310-23. doi: 10.1261/rna.697308. Epub 2007 Dec 14. RNA. 2008. PMID: 18083837 Free PMC article.
References
-
- Agaisse, H., and D. Lereclus. 1994. Structural and functional analysis of the promoter region involved in full expression of the cryIIIA toxin gene of Bacillus thuringiensis. Mol. Microbiol. 13:97-107. - PubMed
-
- Agaisse, H., and D. Lereclus. 1996. STAB-SD: a Shine-Dalgarno sequence in the 5′ untranslated region is a determinant of mRNA stability. Mol. Microbiol. 20:633-643. - PubMed
-
- Alifano, P., F. Rivellini, C. Piscitelli, C. M. Arraiano, C. B. Bruni, and M. S. Carlomagno. 1994. Ribonuclease E provides substrates for ribonuclease P-dependent processing of a polycistronic mRNA. Genes Dev. 8:3021-3031. - PubMed
-
- Allmansberger, R. 1996. Degradation of the Bacillus subtilis xynA transcript is accelerated in response to stress. Mol. Gen. Genet. 251:108-112. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

