Conjugative plasmid transfer in gram-positive bacteria
- PMID: 12794193
- PMCID: PMC156469
- DOI: 10.1128/MMBR.67.2.277-301.2003
Conjugative plasmid transfer in gram-positive bacteria
Abstract
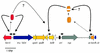
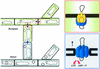
Conjugative transfer of bacterial plasmids is the most efficient way of horizontal gene spread, and it is therefore considered one of the major reasons for the increase in the number of bacteria exhibiting multiple-antibiotic resistance. Thus, conjugation and spread of antibiotic resistance represents a severe problem in antibiotic treatment, especially of immunosuppressed patients and in intensive care units. While conjugation in gram-negative bacteria has been studied in great detail over the last decades, the transfer mechanisms of antibiotic resistance plasmids in gram-positive bacteria remained obscure. In the last few years, the entire nucleotide sequences of several large conjugative plasmids from gram-positive bacteria have been determined. Sequence analyses and data bank comparisons of their putative transfer (tra) regions have revealed significant similarities to tra regions of plasmids from gram-negative bacteria with regard to the respective DNA relaxases and their targets, the origins of transfer (oriT), and putative nucleoside triphosphatases NTP-ases with homologies to type IV secretion systems. In contrast, a single gene encoding a septal DNA translocator protein is involved in plasmid transfer between micelle-forming streptomycetes. Based on these clues, we propose the existence of two fundamentally different plasmid-mediated conjugative mechanisms in gram-positive microorganisms, namely, the mechanism taking place in unicellular gram-positive bacteria, which is functionally similar to that in gram-negative bacteria, and a second type that occurs in multicellular gram-positive bacteria, which seems to be characterized by double-stranded DNA transfer.
Figures








References
-
- An, F. Y., and D. B. Clewell. 1997. The origin of transfer (oriT) of the enterococcal pheromone-responding, cytolysin plasmid pAD1 is located within the repA determinant. Plasmid 37:87-94. - PubMed
-
- Andrup, L., O. Jorgensen, A. Wilcks, L. Smidt, and G. B. Jensen. 1996. Mobilization of “nonmobilizable” plasmids by the aggregation-mediated conjugation system of Bacillus thuringiensis. Plasmid 36:75-85. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

