A data review and re-assessment of ovarian cancer serum proteomic profiling
- PMID: 12795817
- PMCID: PMC165662
- DOI: 10.1186/1471-2105-4-24
A data review and re-assessment of ovarian cancer serum proteomic profiling
Abstract
Background: The early detection of ovarian cancer has the potential to dramatically reduce mortality. Recently, the use of mass spectrometry to develop profiles of patient serum proteins, combined with advanced data mining algorithms has been reported as a promising method to achieve this goal. In this report, we analyze the Ovarian Dataset 8-7-02 downloaded from the Clinical Proteomics Program Databank website, using nonparametric statistics and stepwise discriminant analysis to develop rules to diagnose patients, as well as to understand general patterns in the data that may guide future research.
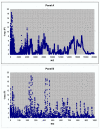
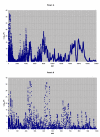
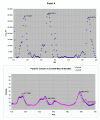
Results: The mass spectrometry serum profiles derived from cancer and controls exhibited numerous statistical differences. For example, use of the Wilcoxon test in comparing the intensity at each of the 15,154 mass to charge (M/Z) values between the cancer and controls, resulted in the detection of 3,591 M/Z values whose intensities differed by a p-value of 10-6 or less. The region containing the M/Z values of greatest statistical difference between cancer and controls occurred at M/Z values less than 500. For example the M/Z values of 2.7921478 and 245.53704 could be used to significantly separate the cancer from control groups. Three other sets of M/Z values were developed using a training set that could distinguish between cancer and control subjects in a test set with 100% sensitivity and specificity.
Conclusion: The ability to discriminate between cancer and control subjects based on the M/Z values of 2.7921478 and 245.53704 reveals the existence of a significant non-biologic experimental bias between these two groups. This bias may invalidate attempts to use this dataset to find patterns of reproducible diagnostic value. To minimize false discovery, results using mass spectrometry and data mining algorithms should be carefully reviewed and benchmarked with routine statistical methods.
Figures

References
-
- Srinivas PR, Verma M, Zhao Y, Srivastava S. Proteomics for cancer biomarker discovery. Clin Chem. 2002;48:1160–1169. - PubMed
-
- Clinical Proteomics Data Bank http://clinicalproteomics.steem.com/ppatterns.php
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

