RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei
- PMID: 12796299
- PMCID: PMC161450
- DOI: 10.1128/EC.2.3.542-551.2003
RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei
Abstract
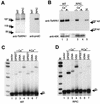
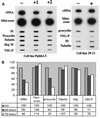
In eukaryotes, RNA polymerase (pol) I exclusively transcribes the large rRNA gene unit (rDNA) and mRNA is synthesized by RNA pol II. The African trypanosome, Trypanosoma brucei, represents an exception to this rule. In this organism, transcription of genes encoding the variant surface glycoprotein (VSG) and the procyclins is resistant to alpha-amanitin, indicating that it is mediated by RNA pol I, while other protein-coding genes are transcribed by RNA pol II. To obtain firm proof for this concept, we generated a T. brucei cell line which exclusively expresses protein C epitope-tagged RNA pol I. Using an anti-protein C immunoaffinity matrix, we specifically depleted RNA pol I from transcriptionally active cell extracts. The depletion of RNA pol I impaired in vitro transcription initiated at the rDNA promoter, the GPEET procyclin gene promoter, and a VSG gene expression site promoter but did not affect transcription from the spliced leader (SL) RNA gene promoter. Fittingly, induction of RNA interference against the RNA pol I largest subunit in insect-form trypanosomes significantly reduced the relative transcriptional efficiency of rDNA, procyclin genes, and VSG expression sites in vivo whereas that of SL RNA, alphabeta-tubulin, and heat shock protein 70 genes was not affected. Our studies unequivocally show that T. brucei harbors a multifunctional RNA pol I which, in addition to transcribing rDNA, transcribes procyclin genes and VSG gene expression sites.
Figures

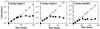


Similar articles
-
Transcription of protein-coding genes in trypanosomes by RNA polymerase I.Annu Rev Microbiol. 1997;51:463-89. doi: 10.1146/annurev.micro.51.1.463. Annu Rev Microbiol. 1997. PMID: 9343357 Review.
-
Alpha-amanitin-resistant transcription units in trypanosomes: a comparison of promoter sequences for a VSG gene expression site and for the ribosomal RNA genes.Nucleic Acids Res. 1991 Oct 11;19(19):5153-8. doi: 10.1093/nar/19.19.5153. Nucleic Acids Res. 1991. PMID: 1923801 Free PMC article.
-
In-vitro competition analysis of procyclin gene and variant surface glycoprotein gene expression site transcription in Trypanosoma brucei.Mol Biochem Parasitol. 2001 Mar;113(1):55-65. doi: 10.1016/s0166-6851(00)00380-7. Mol Biochem Parasitol. 2001. PMID: 11254954
-
Nuclear repositioning of the VSG promoter during developmental silencing in Trypanosoma brucei.J Cell Biol. 2007 Jan 15;176(2):133-9. doi: 10.1083/jcb.200607174. Epub 2007 Jan 8. J Cell Biol. 2007. PMID: 17210949 Free PMC article.
-
Nuclear architecture underlying gene expression in Trypanosoma brucei.Trends Microbiol. 2007 Jun;15(6):263-70. doi: 10.1016/j.tim.2007.04.004. Epub 2007 May 4. Trends Microbiol. 2007. PMID: 17481901 Review.
Cited by
-
The RRM-mediated RNA binding activity in T. brucei RAP1 is essential for VSG monoallelic expression.Nat Commun. 2023 Mar 22;14(1):1576. doi: 10.1038/s41467-023-37307-0. Nat Commun. 2023. PMID: 36949076 Free PMC article.
-
Trypanosoma brucei TIF2 suppresses VSG switching by maintaining subtelomere integrity.Cell Res. 2014 Jul;24(7):870-85. doi: 10.1038/cr.2014.60. Epub 2014 May 9. Cell Res. 2014. PMID: 24810301 Free PMC article.
-
A quorum sensing-independent path to stumpy development in Trypanosoma brucei.PLoS Pathog. 2017 Apr 10;13(4):e1006324. doi: 10.1371/journal.ppat.1006324. eCollection 2017 Apr. PLoS Pathog. 2017. PMID: 28394929 Free PMC article.
-
Dynein Light Chain LC8 Is Required for RNA Polymerase I-Mediated Transcription in Trypanosoma brucei, Facilitating Assembly and Promoter Binding of Class I Transcription Factor A.Mol Cell Biol. 2015 Oct 12;36(1):95-107. doi: 10.1128/MCB.00705-15. Print 2016 Jan 1. Mol Cell Biol. 2015. PMID: 26459761 Free PMC article.
-
An assembly of nuclear bodies associates with the active VSG expression site in African trypanosomes.Nat Commun. 2022 Jan 10;13(1):101. doi: 10.1038/s41467-021-27625-6. Nat Commun. 2022. PMID: 35013170 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

