RBP38, a novel RNA-binding protein from trypanosomatid mitochondria, modulates RNA stability
- PMID: 12796301
- PMCID: PMC161464
- DOI: 10.1128/EC.2.3.560-568.2003
RBP38, a novel RNA-binding protein from trypanosomatid mitochondria, modulates RNA stability
Abstract
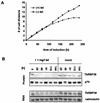
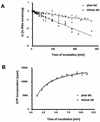
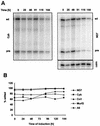
We describe here the isolation and characterization of a novel RNA-binding protein, RBP38, from Leishmania tarentolae mitochondria. This protein does not contain any known RNA-binding motifs and is highly conserved among the trypanosomatids, but no homologues were found in other organisms. Recombinant LtRBP38 binds single and double-stranded (ds) RNA substrates with dissociation constants in the 100 nM range, as determined by fluorescence polarization analysis. Downregulation of expression of the homologous gene, TbRBP38, in procyclic Trypanosoma brucei by using conditional dsRNA interference resulted in 80% reduction of steady-state levels of RNAs transcribed from both maxicircle and minicircle DNA. In organello pulse-chase labeling experiments were used to determine the stability of RNAs in mitochondria that were depleted of TbRBP38. The half-life of metabolically labeled RNA decreased from approximately 160 to approximately 60 min after depletion. In contrast, there was no change in transcriptional activity. These observations suggest a role of RBP38 in stabilizing mitochondrial RNA.
Figures
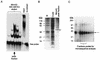




Similar articles
-
A 100-kD complex of two RNA-binding proteins from mitochondria of Leishmania tarentolae catalyzes RNA annealing and interacts with several RNA editing components.RNA. 2003 Jan;9(1):62-76. doi: 10.1261/rna.2134303. RNA. 2003. PMID: 12554877 Free PMC article.
-
Purification, cloning, and expression of two closely related Trypanosoma brucei nucleic acid binding proteins.Mol Biochem Parasitol. 1997 Aug;87(2):145-58. doi: 10.1016/s0166-6851(97)00060-1. Mol Biochem Parasitol. 1997. PMID: 9247926
-
Comparison of the Mitochondrial Genomes and Steady State Transcriptomes of Two Strains of the Trypanosomatid Parasite, Leishmania tarentolae.PLoS Negl Trop Dis. 2015 Jul 23;9(7):e0003841. doi: 10.1371/journal.pntd.0003841. eCollection 2015. PLoS Negl Trop Dis. 2015. PMID: 26204118 Free PMC article.
-
Characterization of two nuclear-encoded protein components of mitochondrial ribonucleoprotein complexes from Leishmania tarentolae.Mol Biochem Parasitol. 1995 Apr;71(1):65-79. doi: 10.1016/0166-6851(95)00023-t. Mol Biochem Parasitol. 1995. PMID: 7630384
-
RNA editing and the mysterious undercover genes of trypanosomatid mitochondria.Trends Biochem Sci. 1988 Aug;13(8):283-4. doi: 10.1016/0968-0004(88)90117-x. Trends Biochem Sci. 1988. PMID: 3154277 Review. No abstract available.
Cited by
-
Naphthalene-based RNA editing inhibitor blocks RNA editing activities and editosome assembly in Trypanosoma brucei.J Biol Chem. 2011 Apr 22;286(16):14178-89. doi: 10.1074/jbc.M110.199646. Epub 2011 Mar 4. J Biol Chem. 2011. PMID: 21378165 Free PMC article.
-
The Trypanosoma cruzi nucleic acid binding protein Tc38 presents changes in the intramitochondrial distribution during the cell cycle.BMC Microbiol. 2009 Feb 11;9:34. doi: 10.1186/1471-2180-9-34. BMC Microbiol. 2009. PMID: 19210781 Free PMC article.
-
TbRGG1, an essential protein involved in kinetoplastid RNA metabolism that is associated with a novel multiprotein complex.RNA. 2008 May;14(5):970-80. doi: 10.1261/rna.888808. Epub 2008 Mar 27. RNA. 2008. PMID: 18369185 Free PMC article.
-
Role of p38 in replication of Trypanosoma brucei kinetoplast DNA.Mol Cell Biol. 2006 Jul;26(14):5382-93. doi: 10.1128/MCB.00369-06. Mol Cell Biol. 2006. PMID: 16809774 Free PMC article.
-
A type III protein arginine methyltransferase from the protozoan parasite Trypanosoma brucei.J Biol Chem. 2009 Apr 24;284(17):11590-600. doi: 10.1074/jbc.M807279200. Epub 2009 Mar 2. J Biol Chem. 2009. PMID: 19254949 Free PMC article.
References
-
- Aphasizhev, R., S. Sbicego, M. Peris, S. H. Jang, I. Aphasizheva, A. M. Simpson, A. Rivlin, and L. Simpson. 2002. Trypanosome mitochondrial 3′-terminal uridylyl transferase (TUTase): the key enzyme in U-insertion/deletion RNA editing. Cell 108:637-648. - PubMed
-
- Blum, B., N. Bakalara, and L. Simpson. 1990. A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 60:189-198. - PubMed
-
- Braly, P., L. Simpson, and F. Kretzer. 1974. Isolation of kinetoplast-mitochondrial complexes from Leishmania tarentolae. J. Protozool. 21:782-790. - PubMed
-
- Brun, R., and Schonenberger. 1979. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 36:289-292. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

