Changing patterns of gene expression in dictyostelium prestalk cell subtypes recognized by in situ hybridization with genes from microarray analyses
- PMID: 12796308
- PMCID: PMC161460
- DOI: 10.1128/EC.2.3.627-637.2003
Changing patterns of gene expression in dictyostelium prestalk cell subtypes recognized by in situ hybridization with genes from microarray analyses
Abstract
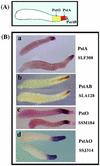
We used microarrays carrying most of the genes that are developmentally regulated in Dictyostelium to discover those that are preferentially expressed in prestalk cells. Prestalk cells are localized at the front of slugs and play crucial roles in morphogenesis and slug migration. Using whole-mount in situ hybridization, we were able to verify 104 prestalk genes. Three of these were found to be expressed only in cells at the very front of slugs, the PstA cell type. Another 10 genes were found to be expressed in the small number of cells that form a central core at the anterior, the PstAB cell type. The rest of the prestalk-specific genes are expressed in PstO cells, which are found immediately posterior to PstA cells but anterior to 80% of the slug that consists of prespore cells. Half of these are also expressed in PstA cells. At later stages of development, the patterns of expression of a considerable number of these prestalk genes changes significantly, allowing us to further subdivide them. Some are expressed at much higher levels during culmination, while others are repressed. These results demonstrate the extremely dynamic nature of cell-type-specific expression in Dictyostelium and further define the changing physiology of the cell types. One of the signals that affect gene expression in PstO cells is the hexaphenone DIF-1. We found that expression of about half of the PstO-specific genes were affected in a mutant that is unable to synthesize DIF-1, while the rest appeared to be DIF independent. These results indicate that differentiation of some aspects of PstO cells can occur in the absence of DIF-1.
Figures







Similar articles
-
The role of DIF-1 signaling in Dictyostelium development.Mol Cell. 2000 Dec;6(6):1509-14. doi: 10.1016/s1097-2765(00)00147-7. Mol Cell. 2000. PMID: 11163223
-
Control of cell type proportioning in Dictyostelium discoideum by differentiation-inducing factor as determined by in situ hybridization.Eukaryot Cell. 2004 Oct;3(5):1241-8. doi: 10.1128/EC.3.5.1241-1248.2004. Eukaryot Cell. 2004. PMID: 15470253 Free PMC article.
-
Regulation of Dictyostelium prestalk-specific gene expression by a SHAQKY family MYB transcription factor.Development. 2006 May;133(9):1715-24. doi: 10.1242/dev.02327. Epub 2006 Mar 29. Development. 2006. PMID: 16571632
-
Interacting signalling pathways regulating prestalk cell differentiation and movement during the morphogenesis of Dictyostelium.Dev Suppl. 1993:1-7. Dev Suppl. 1993. PMID: 8049465 Review.
-
Regulation of Dictyostelium morphogenesis by cAMP-dependent protein kinase.Philos Trans R Soc Lond B Biol Sci. 1993 Jun 29;340(1293):305-13. doi: 10.1098/rstb.1993.0072. Philos Trans R Soc Lond B Biol Sci. 1993. PMID: 8103933 Review.
Cited by
-
Treasure hunt in an amoeba: non-coding RNAs in Dictyostelium discoideum.Curr Genet. 2007 Mar;51(3):141-59. doi: 10.1007/s00294-006-0112-z. Curr Genet. 2007. PMID: 17171561 Review.
-
Genome-wide expression analyses of gene regulation during early development of Dictyostelium discoideum.Eukaryot Cell. 2003 Aug;2(4):664-70. doi: 10.1128/EC.2.4.664-670.2003. Eukaryot Cell. 2003. PMID: 12912885 Free PMC article.
-
GBF-dependent family genes morphologically suppress the partially active Dictyostelium STATa strain.Dev Genes Evol. 2008 Feb;218(2):55-68. doi: 10.1007/s00427-008-0202-7. Epub 2008 Jan 18. Dev Genes Evol. 2008. PMID: 18204858
-
Cell type specificity of a diffusible inducer is determined by a GATA family transcription factor.Development. 2008 May;135(9):1635-45. doi: 10.1242/dev.020883. Epub 2008 Mar 26. Development. 2008. PMID: 18367552 Free PMC article.
-
Mutual suppression between mutations in the Dictyostelium Greenbeard pathway restores wild-type development.BMC Genomics. 2025 Jun 5;26(1):563. doi: 10.1186/s12864-025-11745-0. BMC Genomics. 2025. PMID: 40474076 Free PMC article.
References
-
- Blume, J. E., and H. L. Ennis. 1991. A Dictyostelium discoideum cellulase is a member of a spore germination-specific gene family. J. Biol. Chem. 266:15432-15437. - PubMed
-
- Borth, W., and D. Ratner. 1983. Different synthetic profiles and developmental fates of prespore versus prestalk proteins of Dictyostelium. Differentiation 24:213-219. - PubMed
-
- Bush, J., J. Richardson, and J. Cardelli. 1994. Molecular cloning and characterization of the full-length cDNA encoding the developmentally regulated lysosomal enzyme beta-glucosidase in Dictyostelium discoideum. J. Biol. Chem. 269:1468-1476. - PubMed
-
- Coukell, B., J. Moniakis, and A. Grinberg. 1995. Cloning and expression in Escherichia coli of a cDNA encoding a developmentally regulated Ca(2+)-binding protein from Dictyostelium discoideum. FEBS Lett. 362:342-346. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

