MFE1, a member of the peroxisomal hydroxyacyl coenzyme A dehydrogenase family, affects fatty acid metabolism necessary for morphogenesis in Dictyostelium spp
- PMID: 12796309
- PMCID: PMC161440
- DOI: 10.1128/EC.2.3.638-645.2003
MFE1, a member of the peroxisomal hydroxyacyl coenzyme A dehydrogenase family, affects fatty acid metabolism necessary for morphogenesis in Dictyostelium spp
Abstract
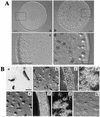
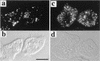
Beta-oxidation of long-chain fatty acids and branched-chain fatty acids is carried out in mammalian peroxisomes by a multifunctional enzyme (MFE) or D-bifunctional protein, with separate domains for hydroxyacyl coenzyme A (CoA) dehydrogenase, enoyl-CoA hydratase, and steroid carrier protein SCP2. We have found that Dictyostelium has a gene, mfeA, encoding MFE1 with homology to the hydroxyacyl-CoA dehydrogenase and SCP2 domains. A separate gene, mfeB, encodes MFE2 with homology to the enoyl-CoA hydratase domain. When grown on a diet of bacteria, Dictyostelium cells in which mfeA is disrupted accumulate excess cyclopropane fatty acids and are unable to develop beyond early aggregation. Axenically grown mutant cells, however, developed into normal fruiting bodies composed of spores and stalk cells. Comparative analysis of whole-cell lipid compositions revealed that bacterially grown mutant cells accumulated cyclopropane fatty acids that remained throughout the developmental stages. Such a persistent accumulation was not detected in wild-type cells or axenically grown mutant cells. Bacterial phosphatidylethanolamine that contains abundant cyclopropane fatty acids inhibited the development of even axenically grown mutant cells, while dipalmitoyl phosphatidylethanolamine did not. These results suggest that MFE1 protects the cells from the increase of the harmful xenobiotic fatty acids incorporated from their diets and optimizes cellular lipid composition for proper development. Hence, we propose that this enzyme plays an irreplaceable role in the survival strategy of Dictyostelium cells to form spores for their efficient dispersal in nature.
Figures




Similar articles
-
Further characterization of the peroxisomal 3-hydroxyacyl-CoA dehydrogenases from rat liver. Relationship between the different dehydrogenases and evidence that fatty acids and the C27 bile acids di- and tri-hydroxycoprostanic acids are metabolized by separate multifunctional proteins.Eur J Biochem. 1996 Sep 15;240(3):660-6. doi: 10.1111/j.1432-1033.1996.0660h.x. Eur J Biochem. 1996. PMID: 8856068
-
Defect in peroxisomal multifunctional enzyme MFE1 affects cAMP relay in Dictyostelium.Dev Growth Differ. 2004 Apr;46(2):195-9. doi: 10.1111/j.1440-169X.2004.00732.x. Dev Growth Differ. 2004. PMID: 15066197
-
Regulation of peroxisome size and number by fatty acid beta -oxidation in the yeast yarrowia lipolytica.J Biol Chem. 2000 Jun 30;275(26):20168-78. doi: 10.1074/jbc.M909285199. J Biol Chem. 2000. PMID: 10787422
-
Peroxisomes and beta-oxidation of long-chain unsaturated carboxylic acids.Scand J Clin Lab Invest Suppl. 1991;204:33-46. doi: 10.3109/00365519109104593. Scand J Clin Lab Invest Suppl. 1991. PMID: 2042025 Review.
-
Peroxisomal lipid degradation via beta- and alpha-oxidation in mammals.Cell Biochem Biophys. 2000;32 Spring:73-87. doi: 10.1385/cbb:32:1-3:73. Cell Biochem Biophys. 2000. PMID: 11330072 Review.
Cited by
-
Fusion and fission, the evolution of sterol carrier protein-2.J Mol Evol. 2006 Mar;62(3):292-306. doi: 10.1007/s00239-005-0086-3. Epub 2006 Feb 20. J Mol Evol. 2006. PMID: 16501878
-
LPIN2 is the phosphatase dominating the penultimate step of neutral lipid biosynthesis in Dictyostelium.MicroPubl Biol. 2024 Oct 23;2024:10.17912/micropub.biology.001296. doi: 10.17912/micropub.biology.001296. eCollection 2024. MicroPubl Biol. 2024. PMID: 39512784 Free PMC article.
-
GBF-dependent family genes morphologically suppress the partially active Dictyostelium STATa strain.Dev Genes Evol. 2008 Feb;218(2):55-68. doi: 10.1007/s00427-008-0202-7. Epub 2008 Jan 18. Dev Genes Evol. 2008. PMID: 18204858
-
Fat-containing cells are eliminated during Dictyostelium development.Biol Open. 2017 Sep 15;6(9):1294-1304. doi: 10.1242/bio.025478. Biol Open. 2017. PMID: 28751309 Free PMC article.
-
Dictyostelium lipid droplets host novel proteins.Eukaryot Cell. 2013 Nov;12(11):1517-29. doi: 10.1128/EC.00182-13. Epub 2013 Sep 13. Eukaryot Cell. 2013. PMID: 24036346 Free PMC article.
References
-
- Adachi, H., T. Hasebe, K. Yoshinaga, T. Ohta, and K. Sutoh. 1994. Isolation of Dictyostelium discoideum cytokinesis mutants by restriction enzyme-mediated integration of the blasticidin S resistance marker. Biochem. Biophys. Res. Commun. 205:1808-1814. - PubMed
-
- Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. - PubMed
-
- Bonner, J. T., and M. K. Slifkin. 1949. A study of the control of differentiation: the proportion of stalk and spore cells in the slime mold Dictyostelium discoideum. Am. J. Bot. 36:727-734.
-
- Breitling, R., Z. Marijanovic, D. Perovic, and J. Adamski. 2001. Evolution of 17β-HSD type 4, a multifunctional protein of β-oxidation. Mol. Cell. Endocrinol. 171:205-210. - PubMed
-
- Escalante, R., and W. F. Loomis. 1995. Whole-mount in situ hybridization of cell-type-specific mRNAs in Dictyostelium. Dev. Biol. 171:262-266. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

