RNA recognition by designed peptide fusion creates "artificial" tRNA synthetase
- PMID: 12796515
- PMCID: PMC164610
- DOI: 10.1073/pnas.1332771100
RNA recognition by designed peptide fusion creates "artificial" tRNA synthetase
Abstract
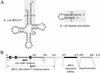
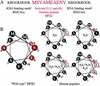
The genetic code was established through aminoacylations of RNA substrates that emerged as tRNAs. The 20 aminoacyl-tRNA synthetases (one for each amino acid) are ancient proteins, the active-site domain of which catalyzes formation of an aminoacyl adenylate that subsequently reacts with the 3' end of bound tRNA. Binding of tRNA depends on idiosyncratic (to the particular synthetase) domains and motifs that are fused to or inserted into the conserved active-site domain. Here we take the domain for synthesis of alanyl adenylate and fuse it to "artificial" peptide sequences (28 aa) that were shown previously to bind to the acceptor arm of tRNAAla. Certain fusions confer aminoacylation activity on tRNAAla and on hairpin microhelices modeled after its acceptor stem. Aminoacylation was sensitive to the presence of a specific G:U base pair known to be a major determinant of tRNAAla identity. Aminoacylation efficiency and specificity also depended on the specific peptide sequence. The results demonstrate that barriers to RNA-specific aminoacylations are low and can be achieved by relatively simple peptide fusions. They also suggest a paradigm for rationally designed specific aminoacylations based on peptide fusions.
Figures



Similar articles
-
Domain-domain communication for tRNA aminoacylation: the importance of covalent connectivity.Biochemistry. 2005 May 17;44(19):7240-9. doi: 10.1021/bi050285y. Biochemistry. 2005. PMID: 15882062
-
Aminoacylation of RNA minihelices: implications for tRNA synthetase structural design and evolution.Crit Rev Biochem Mol Biol. 1993;28(4):309-22. doi: 10.3109/10409239309078438. Crit Rev Biochem Mol Biol. 1993. PMID: 7691478 Review.
-
Assembly of a catalytic unit for RNA microhelix aminoacylation using nonspecific RNA binding domains.Proc Natl Acad Sci U S A. 1999 Oct 26;96(22):12316-21. doi: 10.1073/pnas.96.22.12316. Proc Natl Acad Sci U S A. 1999. PMID: 10535919 Free PMC article.
-
C-terminal zinc-containing peptide required for RNA recognition by a class I tRNA synthetase.Biochemistry. 1996 Apr 2;35(13):4139-45. doi: 10.1021/bi9527810. Biochemistry. 1996. PMID: 8672449
-
Domain-domain communication in aminoacyl-tRNA synthetases.Prog Nucleic Acid Res Mol Biol. 2001;69:317-49. doi: 10.1016/s0079-6603(01)69050-0. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11550797 Review.
Cited by
-
Motif-programmed artificial protein induces apoptosis in several cancer cells by disrupting mitochondria.Cancer Sci. 2008 Feb;99(2):398-406. doi: 10.1111/j.1349-7006.2007.00697.x. Cancer Sci. 2008. PMID: 18271938 Free PMC article.
-
Leucyl-tRNA synthetase from the ancestral bacterium Aquifex aeolicus contains relics of synthetase evolution.EMBO J. 2005 Apr 6;24(7):1430-9. doi: 10.1038/sj.emboj.7600618. Epub 2005 Mar 17. EMBO J. 2005. PMID: 15775966 Free PMC article.
-
Motif programming: a microgene-based method for creating synthetic proteins containing multiple functional motifs.Nucleic Acids Res. 2007;35(6):e38. doi: 10.1093/nar/gkm017. Epub 2007 Feb 7. Nucleic Acids Res. 2007. PMID: 17287291 Free PMC article.
-
Aminoacyl-tRNA Synthetases in the Bacterial World.EcoSal Plus. 2016 May;7(1):10.1128/ecosalplus.ESP-0002-2016. doi: 10.1128/ecosalplus.ESP-0002-2016. EcoSal Plus. 2016. PMID: 27223819 Free PMC article. Review.
-
Single amino acid changes in AspRS reveal alternative routes for expanding its tRNA repertoire in vivo.Nucleic Acids Res. 2004 Aug 2;32(13):4081-9. doi: 10.1093/nar/gkh751. Print 2004. Nucleic Acids Res. 2004. PMID: 15289581 Free PMC article.
References
-
- Ribas de Pouplana, L. & Schimmel, P. (2001) J. Biol. Chem. 276, 6881–6884. - PubMed
-
- Kim, S. H., Suddath, F. L., Quigley, G. J., McPherson, A., Sussman, J. L., Wang, A. H. J., Seeman, N. C. & Rich, A. (1974) Science 185, 435–440. - PubMed
-
- Robertus, J. D., Ladner, J. E., Finch, J. T., Rhodes, D., Brown, R. S., Clark, B. F. C. & Klug, A. (1974) Nature 250, 546–551. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

