High rates of recombination in otitis media isolates of non-typeable Haemophilus influenzae
- PMID: 12797973
- PMCID: PMC2728426
- DOI: 10.1016/s1567-1348(02)00152-1
High rates of recombination in otitis media isolates of non-typeable Haemophilus influenzae
Abstract
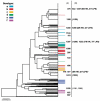
Non-typeable (NT) or capsule-deficient, Haemophilus influenzae (Hi) is a common commensal of the upper respiratory tract of humans and can be pathogenic resulting in diseases such as otitis media, sinusitis and pneumonia. The lipopolysaccharide (LPS) of NTHi is a major virulence factor that displays substantial intra-strain and inter-strain variation of its oligosaccharide structures. To investigate the genetic basis of LPS variation we sequenced internal regions of each of seven genes required for the biosynthesis of either the inner or the outer core oligosaccharide structures. These sequences were obtained from 25 representative NTHi isolates from episodes of otitis media. We found abundant evidence of recombination among LPS genes of NTHi, a finding in marked contrast to previous analyses of biosynthetic genes for capsular polysaccharide, a well-documented virulence factor of Hi. We found mosaic sequences, linkage equilibrium between loci and a lack of congruence between gene trees. These high rates were not confined to LPS genes since evidence for similar amounts of recombination was also found in eight housekeeping genes in a subset of the same 25 isolates. These findings provide a population based foundation for a better understanding of the role of NTHi LPS as a virulence factor and its potential as a candidate vaccine.
Figures


References
-
- Bolduc GR, Bouchet V, Jiang R, Geisselsoder J, Truong-Bolduc Q, Rice PA, Pelton SI, Goldstein R. Variability of OMP P1 and its evaluation as a vaccine candidate against experimental otitis media due to non-typeable Haemophilus influenzae: an unambiguous, multifaceted approach. Infect. Immun. 2000;68:4505–4517. - PMC - PubMed
-
- Drouin G, Prat F, Ell M, Clarke GD. Detecting and characterizing gene conversions between multigene family members. Mol. Biol. Evol. 1999;16:1369–1390. - PubMed
-
- Enright MC, Spratt BG. Multilocus sequence typing. Trends Microbiol. 1999;7:482–487. - PubMed
-
- Feil EJ, Holmes EC, Bessen DE, Chan MS, Day NP, Enright MC, Goldstein R, Hood DW, Kalia A, Moore CE, Zhou J, Spratt BG. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. U.S.A. 2001;98:182–187. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

