The C-terminal domain of Escherichia coli MutY is involved in DNA binding and glycosylase activities
- PMID: 12799430
- PMCID: PMC162338
- DOI: 10.1093/nar/gkg434
The C-terminal domain of Escherichia coli MutY is involved in DNA binding and glycosylase activities
Abstract
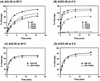
Escherichia coli MutY is an adenine and a weak guanine DNA glycosylase involved in reducing mutagenic effects of 7,8-dihydro-8-oxo-guanine (8-oxoG). The C-terminal domain of MutY is required for 8-oxoG recognition and is critical for mutation avoidance of oxidative damage. To determine which residues of this domain are involved in 8-oxoG recognition, we constructed four MutY mutants based on similarities to MutT, which hydrolyzes specifically 8-oxo-dGTP to 8-oxo-dGMP. F294A-MutY has a slightly reduced binding affinity to A/G mismatch but has a severe defect in A/8-oxoG binding at 20 degrees C. The catalytic activity of F294A-MutY is much weaker than that of the wild-type MutY. The DNA binding activity of R249A-MutY is comparable to that of the wild-type enzyme but the catalytic activity is reduced with both A/G and A/8-oxoG mismatches. The biochemical activities of F261A-MutY are nearly similar to those of the wild-type enzyme. The solubility of P262A-MutY was improved as a fusion protein containing streptococcal protein G (GB1 domain) at its N-terminus. The binding of GB1-P262A-MutY with both A/G and A/8-oxoG mismatches are slightly weaker than those of the wild-type protein. The catalytic activity of GB1-P262A-MutY is weaker than that of the wild-type enzyme at lower enzyme concentrations. Importantly, all four mutants can complement mutY mutants in vivo when expressed at high levels; however, F294A, R249A and P262A, but not F261A, are partially defective in vivo when they are expressed at low levels. These results strongly support that the C-terminal domain of MutY is involved not only in 8-oxoG recognition, but also affects the binding and catalytic activities toward A/G mismatches.
Figures





References
-
- Ames B.N. and Shigenaga,M.K. (1993) Oxidants are a major contributor to cancer and aging. In Halliwell,B. and Aruoma,O. (eds), DNA and Free Radicals. Ellis Horwood, New York, NY, pp. 1–18.
-
- Halliwell B. and Gutteridge,J.M.C. (1989) Free radicals in Biology and Medicine. Oxford University Press, New York.
-
- Tchou J. and Grollman,A.P. (1993) Repair of DNA containing the oxidatively-damaged base 8-hydroxyguanine. Mutat. Res., 299, 277–287. - PubMed

