The Upf-dependent decay of wild-type PPR1 mRNA depends on its 5'-UTR and first 92 ORF nucleotides
- PMID: 12799443
- PMCID: PMC162334
- DOI: 10.1093/nar/gkg430
The Upf-dependent decay of wild-type PPR1 mRNA depends on its 5'-UTR and first 92 ORF nucleotides
Abstract
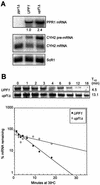
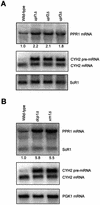
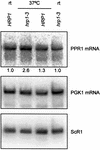
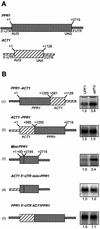
mRNAs containing premature translation termination codons (nonsense mRNAs) are targeted for deadenylation-independent degradation in a mechanism that depends on Upf1p, Upf2p and Upf3p. This decay pathway is often called nonsense- mediated mRNA decay (NMD). Nonsense mRNAs are decapped by Dcp1p and then degraded 5' to 3' by Xrn1p. In the yeast Saccharomyces cerevisiae, a significant number of wild-type mRNAs accumulate in upf mutants. Wild-type PPR1 mRNA is one of these mRNAs. Here we show that PPR1 mRNA degradation depends on the Upf proteins, Dcp1p, Xrn1p and Hrp1p. We have mapped an Upf1p-dependent destabilizing element to a region located within the 5'-UTR and the first 92 bases of the PPR1 ORF. This element targets PPR1 mRNA for Upf-dependent decay by a novel mechanism.
Figures

Similar articles
-
Initiation-mediated mRNA decay in yeast affects heat-shock mRNAs, and works through decapping and 5'-to-3' hydrolysis.Nucleic Acids Res. 2003 Jul 15;31(14):4006-16. doi: 10.1093/nar/gkg474. Nucleic Acids Res. 2003. PMID: 12853617 Free PMC article.
-
Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs.Mol Cell Biol. 2001 Mar;21(5):1515-30. doi: 10.1128/MCB.21.5.1515-1530.2001. Mol Cell Biol. 2001. PMID: 11238889 Free PMC article.
-
Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5' to 3' mRNA decay pathways in yeast.Mol Cell. 2003 Dec;12(6):1439-52. doi: 10.1016/s1097-2765(03)00446-5. Mol Cell. 2003. PMID: 14690598
-
Upf proteins: highly conserved factors involved in nonsense mRNA mediated decay.Mol Biol Rep. 2018 Feb;45(1):39-55. doi: 10.1007/s11033-017-4139-7. Epub 2017 Dec 27. Mol Biol Rep. 2018. PMID: 29282598 Review.
-
mRNA decay: x (XRN1) marks the spot.Mol Cell. 2003 May;11(5):1126-8. doi: 10.1016/s1097-2765(03)00198-9. Mol Cell. 2003. PMID: 12769838 Review.
Cited by
-
Role for Upf2p phosphorylation in Saccharomyces cerevisiae nonsense-mediated mRNA decay.Mol Cell Biol. 2006 May;26(9):3390-400. doi: 10.1128/MCB.26.9.3390-3400.2006. Mol Cell Biol. 2006. PMID: 16611983 Free PMC article.
-
Long 3'-UTRs target wild-type mRNAs for nonsense-mediated mRNA decay in Saccharomyces cerevisiae.Nucleic Acids Res. 2009 May;37(9):2771-8. doi: 10.1093/nar/gkp146. Epub 2009 Mar 6. Nucleic Acids Res. 2009. PMID: 19270062 Free PMC article.
-
Functional polymorphism in the 5'-UTR of CR2 is associated with susceptibility to nasopharyngeal carcinoma.Oncol Rep. 2013 Jul;30(1):11-6. doi: 10.3892/or.2013.2421. Epub 2013 Apr 23. Oncol Rep. 2013. PMID: 23612877 Free PMC article.
-
Gene set coregulated by the Saccharomyces cerevisiae nonsense-mediated mRNA decay pathway.Eukaryot Cell. 2005 Dec;4(12):2066-77. doi: 10.1128/EC.4.12.2066-2077.2005. Eukaryot Cell. 2005. PMID: 16339724 Free PMC article.
-
Candida albicans Tup1 is involved in farnesol-mediated inhibition of filamentous-growth induction.Eukaryot Cell. 2008 Jun;7(6):980-7. doi: 10.1128/EC.00357-07. Epub 2008 Apr 18. Eukaryot Cell. 2008. PMID: 18424510 Free PMC article.
References
-
- Maquat L.E. (2000) Nonsense-mediated RNA decay in mammalian cells: a splicing-dependent means to down-regulate the levels of mRNAs that prematurely terminate translation. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 849–869.
-
- Gonzalez C.I., Bhattacharya,A., Wang,W. and Peltz,S.W. (2001) Nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Gene, 274, 15–25. - PubMed
-
- Leeds P., Peltz,S.W., Jacobson,A. and Culbertson,M.R. (1991) The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev., 5, 2303–2314. - PubMed
-
- Cui Y., Hagan,K.W., Zhang,S. and Peltz,S.W. (1995) Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev., 9, 423–436. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

