Characterization of the 2'-5'-oligoadenylate synthetase ubiquitin-like family
- PMID: 12799444
- PMCID: PMC162331
- DOI: 10.1093/nar/gkg427
Characterization of the 2'-5'-oligoadenylate synthetase ubiquitin-like family
Abstract
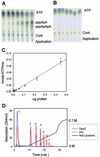
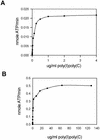
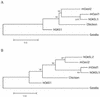
The interferon-induced 2'-5'-oligoadenylate synthetases (OAS) are important for the antiviral activity of interferons. The human and murine OAS gene families each contain four genes: OAS1, OAS2, OAS3 and OASL, all having one or more conserved OAS units composed of five translated exons. The OASL gene has both an OAS unit and a C-terminus of two ubiquitin-like repeats. In this study, we demonstrate that murine Oasl1 protein is inactive while murine Oasl2 is active as an OAS. Further more, murine Oasl2 requires double-stranded RNA as co-factor. The affinity of murine Oasl2 for the double-stranded RNA activator is higher than that of human OAS1 (p42 isoform). We propose a model for the evolutionary origin of the murine Oasl1 and Oasl2 genes. The identification of a human orthologue (hOASL2) to the murine Oasl2 gene establishes that the OASL gene was duplicated prior to the radiation of the rodent and primate groups. We suggest that murine Oasl2, which has both enzymatic activity and a ubiquitin-like domain, is a functional intermediate between the active OAS species and the inactive human OASL1/murine Oasl1 proteins. In addition, we propose that murine Oasl1 appears to have gained a hitherto uncharacterized function independent of 2'-5'-linked oligoadenylate synthesis.
Figures



References
-
- Stark G.R., Kerr,I.M., Williams,B.R., Silverman,R.H. and Schreiber,R.D. (1998) How cells respond to interferons. Annu. Rev. Biochem., 67, 227–264. - PubMed
-
- Roberts W.K., Hovanessian,A., Brown,R.E., Clemens,M.J. and Kerr,I.M (1976) Interferon-mediated protein kinase and low-molecular-weight inhibitor of protein synthesis. Nature, 264, 477–480. - PubMed
-
- Hovanessian A., Brown,R.E. and Kerr,I.M. (1977) Synthesis of low molecular weight inhibitor of protein synthesis with enzyme from interferon-treated cells. Nature, 268, 537–540. - PubMed
-
- Chebath J., Benech,P., Revel,M. and Vigneron,M. (1987) Constitutive expression of (2′–5′) oligo A synthetase confers resistance to picornavirus infection. Nature, 330, 587–588. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

