Alternatively spliced isoforms of the human constitutive androstane receptor
- PMID: 12799447
- PMCID: PMC162252
- DOI: 10.1093/nar/gkg419
Alternatively spliced isoforms of the human constitutive androstane receptor
Abstract
The nuclear receptor CAR (NR1I3) regulates transcription of genes encoding xenobiotic- and steroid-metabolizing enzymes. Regulatory processes that are mediated by CAR are modulated by a structurally diverse array of chemicals including common pharmaceutical and environmental agents. Here we describe four in-frame splice variants of the human CAR receptor gene. The variant mRNA splice transcripts were expressed in all human livers evaluated. Molecular modeling of the splice variant proteins predicts that the structural effects are localized within the receptor's ligand-binding domain. Assays to assess function indicate that the variant proteins, when compared with the reference protein isoform, exhibit compromised activities with respect to DNA binding, transcriptional activation and coactivator recruitment.
Figures





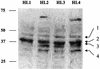



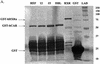

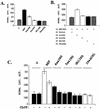
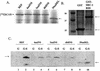
References
-
- Goodwin B., Hodgson,E., D’Costa,D.J., Robertson,G.R. and Liddle,C. (2002) Transcriptional regulation of the human CYP3A4 gene by the constitutive androstane receptor. Mol. Pharmacol., 62, 359–365. - PubMed
-
- Kast H.R., Goodwin,B., Tarr,P.T., Jones,S.A., Anisfeld,A.M., Stoltz,C.M., Tontonoz,P., Kliewer,S., Willson,T.M. and Edwards,P.A. (2002) Regulation of multidrug resistance-associated protein 2 (ABCC2) by the nuclear receptors pregnane X receptor, farnesoid X-activated receptor and constitutive androstane receptor. J. Biol. Chem., 277, 2908–2915. - PubMed
-
- Sueyoshi T. and Negishi,M. (2001) Phenobarbital response elements of cytochrome P450 genes and nuclear receptors. Annu. Rev. Pharmacol. Toxicol., 41, 123–143. - PubMed
-
- Sugatani J., Kojima,H., Ueda,A., Kakizaki,S., Yoshinari,K., Gong,Q.H., Owens,I.S., Negishi,M. and Sueyoshi,T. (2001) The phenobarbital response enhancer module in the human bilirubin UDP-glucuronosyltransferase UGT1A1 gene and regulation by the nuclear receptor CAR. Hepatology, 33, 1232–1238. - PubMed
-
- Xiong H., Yoshinari,K., Brouwer,K.L. and Negishi,M. (2002) Role of constitutive androstane receptor in the in vivo induction of Mrp3 and CYP2B1/2 by phenobarbital. Drug Metab. Disposition, 30, 918–923. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

