Stimulation of Tat-independent transcriptional processivity from the HIV-1 LTR promoter by matrix attachment regions
- PMID: 12799452
- PMCID: PMC162244
- DOI: 10.1093/nar/gkg410
Stimulation of Tat-independent transcriptional processivity from the HIV-1 LTR promoter by matrix attachment regions
Abstract
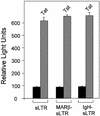
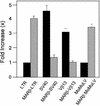
The chromatin environment and the sites of integration in the host genome are critical determinants of human immunodeficiency virus (HIV) transcription and replication. Depending on the chromosomal location of provirus integration within the genome, HIV-1 long terminal repeat (LTR)-mediated transcription may vary from 0- to 70-fold. Cis-elements such as topoisomerase II cleavage sites, Alu repeats and matrix attachment regions (MARs) are thought to be targets for retroviral integration. Here we show that a novel MAR sequence from the T-cell receptor beta locus (MARbeta) and the IgH MAR mediate transcriptional augmentation when placed upstream of the HIV-1 LTR promoter. The effect of transcriptional augmentation is seen in both transient and stable transfection, indicating its effect even upon integration in the genome. MAR-mediated transcriptional elevation is independent of Tat, and occurs synergistically in the presence of Tat. Further, we show that MAR-mediated transcriptional elevation is specific to the HIV-1 LTR and the Moloney murine leukemia virus LTR promoter. In a transient transfection assay using over-expressed IkappaB, the inhibitor of NF-kappaB, we show that MAR-induced processive transcription is NF-kappaB dependent, signifying the role of local enhancers within the LTR promoter. Furthermore, by RNase protection experiments using proximal and distal probes, we show that MAR-mediated transcriptional upregulation is more prominent at the distal rather than the proximal end, thus indicating the potential role of MARs in promoting elongation.
Figures






Similar articles
-
Distinct transcriptional pathways of TAR-dependent and TAR-independent human immunodeficiency virus type-1 transactivation by Tat.Virology. 1997 Aug 18;235(1):48-64. doi: 10.1006/viro.1997.8672. Virology. 1997. PMID: 9300036
-
Semen Exosomes Promote Transcriptional Silencing of HIV-1 by Disrupting NF-κB/Sp1/Tat Circuitry.J Virol. 2018 Oct 12;92(21):e00731-18. doi: 10.1128/JVI.00731-18. Print 2018 Nov 1. J Virol. 2018. PMID: 30111566 Free PMC article.
-
Inhibition by interferon of herpes simplex virus type 1-activated transcription of tat-defective provirus.Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9573-7. doi: 10.1073/pnas.88.21.9573. Proc Natl Acad Sci U S A. 1991. PMID: 1719535 Free PMC article.
-
Multiple modes of transcriptional regulation by the HIV-1 Tat transactivator.IUBMB Life. 2001 Mar;51(3):175-81. doi: 10.1080/152165401753544241. IUBMB Life. 2001. PMID: 11547919 Review.
-
HIV Tat, its TARgets and the control of viral gene expression.FEMS Microbiol Lett. 2003 Mar 14;220(1):57-65. doi: 10.1016/S0378-1097(03)00067-3. FEMS Microbiol Lett. 2003. PMID: 12644228 Review.
Cited by
-
Anchoring the genome.Genome Biol. 2008 Jan 22;9(1):201. doi: 10.1186/gb-2008-9-1-201. Genome Biol. 2008. PMID: 18226181 Free PMC article. Review.
-
SMAR1 and Cux/CDP modulate chromatin and act as negative regulators of the TCRbeta enhancer (Ebeta).Nucleic Acids Res. 2004 Sep 15;32(16):4862-75. doi: 10.1093/nar/gkh807. Print 2004. Nucleic Acids Res. 2004. PMID: 15371550 Free PMC article.
-
Tat inhibition by didehydro-Cortistatin A promotes heterochromatin formation at the HIV-1 long terminal repeat.Epigenetics Chromatin. 2019 Apr 16;12(1):23. doi: 10.1186/s13072-019-0267-8. Epigenetics Chromatin. 2019. PMID: 30992052 Free PMC article.
-
Global regulator SATB1 recruits beta-catenin and regulates T(H)2 differentiation in Wnt-dependent manner.PLoS Biol. 2010 Jan 26;8(1):e1000296. doi: 10.1371/journal.pbio.1000296. PLoS Biol. 2010. PMID: 20126258 Free PMC article.
References
-
- Sodroski J.G., Rosen,C.A., Wong-Stall,F., Salahuddinn,S.Z., Popovic,M., Arya,S., Gallo,R.C. and Hasteline,W.A. (1985) Trans-acting transcriptional regulation of human T-cell leukemia virus type III LTR. Science, 227, 171–173. - PubMed
-
- Emerman M. and Malim,M. (1998) HIV regulatory/accessory genes; keys to unraveling viral and host cell biology. Science, 280, 1880–1884. - PubMed
-
- Wei P., Garber,M.E., Fang,S.M., Fischer,W.H. and Jones,K.A. (1998) A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop specific binding to TAR RNA. Cell, 92, 451–462. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

