Comparative sequencing of human and chimpanzee MHC class I regions unveils insertions/deletions as the major path to genomic divergence
- PMID: 12799463
- PMCID: PMC164652
- DOI: 10.1073/pnas.1230533100
Comparative sequencing of human and chimpanzee MHC class I regions unveils insertions/deletions as the major path to genomic divergence
Abstract
Despite their high degree of genomic similarity, reminiscent of their relatively recent separation from each other ( approximately 6 million years ago), the molecular basis of traits unique to humans vs. their closest relative, the chimpanzee, is largely unknown. This report describes a large-scale single-contig comparison between human and chimpanzee genomes via the sequence analysis of almost one-half of the immunologically critical MHC. This 1,750,601-bp stretch of DNA, which encompasses the entire class I along with the telomeric part of the MHC class III regions, corresponds to an orthologous 1,870,955 bp of the human HLA region. Sequence analysis confirms the existence of a high degree of sequence similarity between the two species. However, and importantly, this 98.6% sequence identity drops to only 86.7% taking into account the multiple insertions/deletions (indels) dispersed throughout the region. This is functionally exemplified by a large deletion of 95 kb between the virtual locations of human MICA and MICB genes, which results in a single hybrid chimpanzee MIC gene, in a segment of the MHC genetically linked to species-specific handling of several viral infections (HIV/SIV, hepatitis B and C) as well as susceptibility to various autoimmune diseases. Finally, if generalized, these data suggest that evolution may have used the mechanistically more drastic indels instead of the more subtle single-nucleotide substitutions for shaping the recently emerged primate species.
Figures


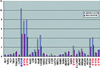


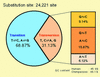
References
-
- Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860-921. - PubMed
-
- Fujiyama, A., Watanabe, H., Toyoda, A., Taylor, T. D., Itoh, T., Tsai, S. F., Park, H. S., Yaspo, M. L., Lehrach, H., Chen, Z., et al. (2002) Science 295, 131-134. - PubMed
-
- Varki, A. (2000) Genome Res. 10, 1065-1070. - PubMed
-
- Koopman, G., Haaksma, A. G., ten Velden, J., Hack, C. E. & Heeney, J. L. (1999) AIDS Res. Hum. Retroviruses 15, 365-373. - PubMed
-
- Dienes, H. P., Purcell, R. H., Popper, H. & Ponzetto, A. (1990) J. Hepatol. 10, 77-84. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

