Suppressed catalytic activity of base excision repair enzymes on rotationally positioned uracil in nucleosomes
- PMID: 12799467
- PMCID: PMC164609
- DOI: 10.1073/pnas.1330328100
Suppressed catalytic activity of base excision repair enzymes on rotationally positioned uracil in nucleosomes
Abstract
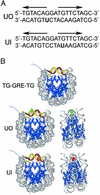
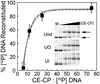
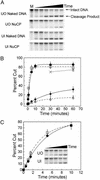
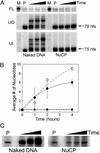
The majority of DNA in eukaryotic cells exists in the highly condensed structural hierarchy of chromatin, which presents a challenge to DNA repair enzymes in that recognition, incision, and restoration of the original sequence at most sites must take place within these structural constraints. To test base excision repair (BER) activities on chromatin substrates, an in vitro system was developed that uses human uracil DNA glycosylase (UDG), apyrimidinic/apurinic endonuclease (APE), and DNA polymerase beta (pol beta) on homogeneously damaged, rotationally positioned DNA in nucleosomes. We find that UDG and APE carry out their combined catalytic activities with reduced efficiency on nucleosome substrates ( approximately 10% of that on naked DNA). Furthermore, these enzymes distinguish between two different rotational settings of the lesion on the histone surface, showing a 2- to 3-fold difference in activity between uracil facing "toward" and "away from" the histones. However, UDG and APE will digest such substrates to completion in a concentration-dependent manner. Conversely, the synthesis activity of pol beta is inhibited completely by nucleosome substrates and is independent of enzyme concentration. These results suggest that the first two steps of BER, UDG and APE, may occur "unassisted" in chromatin, whereas downstream factors in this pathway (i.e., pol beta) may require nucleosome remodeling for efficient DNA BER in at least some regions of chromatin in eukaryotic cells.
Figures
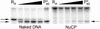
References
-
- Hoeijmakers, J. H. J. (2001) Nature 411, 366–374. - PubMed
-
- Wood, R. D., Mitchell, M., Sgouros, J. & Lindahl, T. (2001) Science 291, 1284–1289. - PubMed
-
- Hanawalt, P. C. (1998) Mutat. Res. 400, 117–125. - PubMed
-
- Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. (1997) Nature 389, 251–260. - PubMed
-
- Wolffe, A. P. (1998) Chromatin: Structure and Function (Academic, San Diego).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

